Abstract
MicroRNAs (miRNAs) are endogenously initiated, small non-coding RNAs and typically regulate the expression of mRNAs in post transcriptional level either via translational repression or mRNA degradation. Aberrant expression of miRNAs is observed in diverse disease and altered physiological states. Recently, it has been revealed that miRNAs are not only present in cells but also in extracellular milieu especially in different bio-fluids including blood plasma, follicular fluid and even in cell culture media. Such extracellular miRNAs (ECmiRNAs) are remarkably stable in the extracellular harsh environment with the presence of high RNAse activity. Although the precise mechanisms of release of cellular miRNAs to extracellular environment remain largely unknown, recent studies suggest that the expression of these ECmiRNAs can be associated with patho-physiological condition of an organism. Moreover, these ECmiRNAs may deliver to the recipient cells via certain pathways where they can regulate translational activity of target genes. This review will discuss the nature and stability of ECmiRNAs along with their release mechanisms. Furthermore, based on recent evidences, it also summarizes the possible function of these ECmiRNAs in distant cell-to-cell communication and the difficulties we may face during ECmiRNA research.
Keywords
Extracellular miRNA ; Circulating miRNA ; Exosomes ; Biomarker
MicroRNA (miRNA) and Their Biogenesis
miRNAs are mostly studied small non-coding RNAs, which are typically 18–24 nucleotides long and are considered as one of the major post-transcriptional regulators of gene expression. This process is accomplished through binding of miRNA to their target mRNAs by base-pairing and subsequently inducing either translational repression or mRNA destabilization. They are estimated to comprise 1–5% of animal genes, and thousands of miRNAs can be encoded by a genome at a time (Hossain et al., 2012 ). Bioinformatics analysis estimated that more than 60% of mRNAs in mammalian genome can be targeted by single miRNA. Because of their broader targeting characteristics, miRNAs are expected to be involved in most biological pathways and cellular processes including cell proliferation, apoptosis, cellular development and cellular signaling. Upon its discovery in Caenorhabditis elegans in the early 1990s ( Lee et al., 1993 ), since then, miRNA has been identified in a wide range of biological pathways of different organisms, ranging from single-cell algae to multi-cellular mammalians, indicating their function is an ancient and critical cellular regulatory mechanism. According to miRbase (version) there are more than 2000 known human miRNAs that have been identified to influence gene expression and associated with pathological conditions.
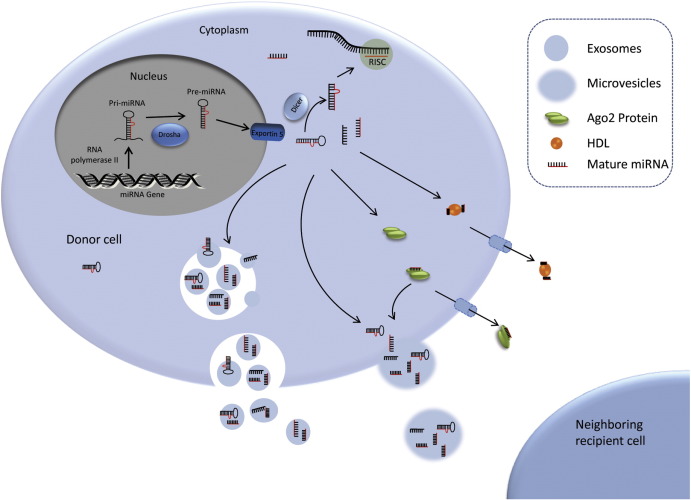
The biogenesis of miRNAs is tightly regulated spatio-temporal process, and any deviation is associated with several diseases. The cellular process of miRNA biogenesis involves both nuclear and cytoplasmic processes (Lee et al., 2002 ). MiRNAs originate from large primary (pri) and precursor (pre) transcripts which undergo successive multi-steps of processing until they reach their mature and functional form (Fig. 1 ). All miRNAs are transcribed by RNA polymerase II from chromosomal DNA (intergenic or intragenic region) into primary transcripts of 1–3 kb long known as pri-miRNAs (Borchert et al ., 2006 ; Garzon et al ., 2010 ; Lee et al ., 2004 ). A microprocessor complex consists of Drosha and DGCR8 further processes the resulting pri-miRNAs in to a ∼ 70 nucleotide long double stranded stem-loop hair-pin like structure called precursor miRNA (pre-miRNA) (Landthaler et al ., 2004 ; Lee et al ., 2003 ). These excited pre-miRNA hairpins are actively transported from nucleus to the cytoplasm by Exportin-5 complexed with Ran-GTPase (Yi et al., 2003 ) where they further cleaved into ~ 22 bp miRNA/miRNA* duplexes by Dicer/TRBP enzyme complex (Chendrimada et al ., 2005 ; Zhang et al ., 2002 ). In the final step, miRNA/miRNA* duplexes separate from each other and subsequently one strand (guide strand) incorporated with Argonaute (AGO) protein of RNA-induced silencing complex (RISC) and forms miRISC whereas the other strand (passenger strand) is supposed to be degraded normally in the cytoplasm (Ender and Meister, 2010 ; Okamura et al ., 2004 ). This mature miRNA strand incorporated with RISC sequence specifically binds to their complementary mRNAs, promoting their degradation or translational inhibit (Fig. 1 ).
|
|
|
Fig. 1. miRNA biogenesis and release of miRNAs in the extracellular environment. Pri-miRNAs are transcribed by RNA polymerase II and later processed by Drosha to Pre-miRNA. Exportin5 transfer these Pre-miRNAs from nucleus to cytoplasm where Dicer processed them into mature miRNAs. Mature miRNAs can be selectively incorporated into the exosomes or coupled with Ago2 protein and released in to extracellular milieu. Alternatively, they can be enwrapped with microvesicles or attached to HDL and later released to extracellular environment. |
Extracellular miRNA (ECmiRNA)
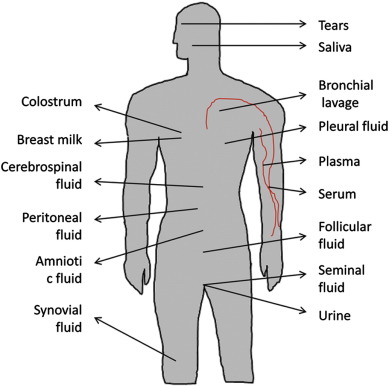
While majority of miRNAs are detected in cellular microenvironment, surprisingly a handful number of miRNAs, commonly known as circulating miRNA or extracellular miRNA, have also been detected in extracellular environment, including different biological fluids (Fig. 2 ) and cell culture media (Hunter et al ., 2008 ; Mitchell et al ., 2008 ; Valadi et al ., 2007 ). Recent studies showed that miRNAs are not only present in serum (Chen et al ., 2008 ; Noferesti et al ., 2015 ) or plasma (Arroyo et al ., 2011 ; Chim et al ., 2008 ) but also in different extracellular bio-fluids including saliva, tears, urine, breast milk, colostrum, peritoneal fluid, cerebrospinal fluid, bronchial lavage and seminal fluid (Weber et al., 2010) and follicular fluid (da Silveira et al ., 2012 ; Sang et al ., 2013 ; Sohel et al ., 2013 ). Moreover, the expression profile of ECmiRNAs from different types of bio-fluids in relation to different pathophysiological conditions shows a specific pattern which indicating that extra-cellular miRNAs may not be passively released from the necrotic or injured cells rather selectively released from the cells (Mar-Aguilar et al ., 2013 ; Noferesti et al ., 2015 ).
|
|
|
Fig. 2. ECmiRNAs found in different bio-fluids so far. ECmiRNAs have been detected in almost all biological fluids including blood serum/plasma, cerebrospinal fluids, peritoneal fluid, amniotic fluid synovial fluid, follicular fluid, breast milk, colostrum, tears and urine. Interestingly, the expression pattern of some of miRNAs in these fluids directly reflect the patho-physiological condition of an organism. |
Unique Characteristics of ECmiRNAs
In contrast to cellular miRNAs and other RNA species, which are degraded in extracellular environment within few seconds, ECmiRNAs are remarkably stable and can survive under unfavorable conditions for long time. When synthetic miRNAs spiked into human plasma, it shows rapid degradation (within few seconds). While denaturing solution inactivated RNase activity in plasma or serum, the exogenous miRNAs get released from degradation (Mitchell et al., 2008 ). Thus, it is clear that synthetic miRNA species are vulnerable and susceptible to quick degradation in plasma or any other bio-fluids, whereas ECmiRNAs are resistant to high endogenous RNase activity, suggesting that these miRNAs are adopting some protective mechanisms to bypass high RNase activity in the extracellular environment. Some reports showed that ECmiRNAs in body fluids remain stable even they subjected to harsh conditions like boiling, high or low pH, prolonged storage time and multiple freeze–thaw cycles while most of cellular RNAs were degraded quickly (Gilad et al ., 2008 ; Taylor and Gercel-Taylor, 2008 ). In addition, other studies demonstrated that miRNAs in serum are still detectable and gives specific expression pattern in quantitative PCR (qPCR) after being subjected to incubation at room temperature for 24 h (Mitchell et al., 2008 ) and maximum 10 freeze–thaw cycle (Chen et al., 2008 ). Furthermore, ECmiRNAs present in serum are resistant against RNase A compared to other endogenous RNA species such as 28s rRNA, 18s rRNA, β-actin, GAPDH and U6 (Chen et al., 2008 ). Most of these ECmiRNAs derived from serum showed considerable level of expression after 3 h or even overnight incubation with RNase-A, however following the same RNase-A treatments all large RNAs were degraded (Chen et al., 2008 ).
The mechanism underlying the remarkable stability of ECmiRNAs in the RNase-rich environment of blood and other bio-fluids are not well understood. Many hypotheses have been proposed to explain the possible mechanisms through which RNAs and miRNAs are released and protected from endogenous RNase activity in circulation. One of the earliest theories suggested that RNAs might be conjugated with protein which would later protect them from both DNase and RNase activity (Sisco, 2001 ). However, later on it has been shown that RNA species present in plasma are protected from degradation probably due to inclusion in lipid or lipoprotein complexes, not by binding with DNA (El-Hefnawy et al., 2004 ). Another hypothesis is that miRNAs are wrapped with membrane vesicles (exosomes, microparticles and apoptotic bodies) that shad miRNAs in extracellular environment and protect them from RNase activity. On the other, some studies have shown that, after isolating exosome/microvesicles using high-speed ultracentrifugation from culture media (Turchinovich et al., 2011 ) or serum (Noferesti et al., 2015 ) or follicular fluid (Sohel et al., 2013 ), a handful number of miRNAs are still detectable in the microvesicles free fraction, suggested that the presence of non-vesicle associated miRNA (might be miRNA-protein or miRNA-lipid/lipoprotein complexes) in extra-cellular fluid.
Possible Ways to Release ECmiRNAs
Although several studies confirmed the presence of ECmiRNAs in various bio-fluids, the mechanisms which are responsible for the release of these miRNAs in extracellular environment remained an unsolved puzzle for quite a long period. There are arguments and theories to explain how these miRNAs released in extracellular body fluids and serves cell–cell communications. One of the earliest assumptions of miRNA release and remarkable stability in extracellular environment is they might be protected by enwrapping into membrane vesicles (i.e. exosomes, microvesicles). This theory emerged after Valadi et al. showed exosomes can carry a functional miRNA population and Hunter et al. detected miRNAs in peripheral blood microvesicles (Hunter et al., 2008 ). These two findings led to a revolutionary theory — existence and transportation of miRNAs can be mediated through vesicles. Later on, increasing number of studies have demonstrated that the transfer of protein, mRNA and miRNA in different body fluids can be mediated via exosomes, microvesicles and apoptotic bodies that are released from a variety of cell types to modulate cell proliferation/apoptosis, angiogenesis, tumor cell invasion and cell–cell communication. In 2011, the theory “vesicle encapsulated existence and transport of miRNA” was hugely challenged when two independent studies showed that 90% ECmiRNAs are associated with Ago protein family in both blood plasma and cell culture media (Arroyo et al ., 2011 ; Turchinovich et al ., 2011 ). However, ECmiRNAs coupled with Ago protein family found to be non-specific product resulting from physiological activity and death of cells (Turchinovich et al., 2012 ). Since then, increasing number of studies have consistently showed that ECmiRNAs can be released and shaded from extracellular harsh environment through a) exosomes b) microvesicles c) apoptotic bodies d) high density lipoprotein (HDL) and e) AGO protein complex. The release mechanisms of ECmiRNAs are graphically presented in Fig. 1 .
Release and Transport of ECmiRNA Through Exosomes
Exosomes are homologous membrane bound small vesicles (50–90 nm) of endosomal origin (Camussi et al ., 2010 ; Cheng et al ., 2014 ) and are present in almost all biological fluids. The main components of exosome membrane are lipid and protein which enriched with lipid rafts (Mathivanan et al., 2010 ). Exosomes are formed by internalization of the cell membrane to produce endosomes. Invagination of the membrane of endosomes results several intraluminal vesicles, these organelles are known as multivesicular bodies (Urbé et al., 2003 ). Budding of endosomes occurs in response to different cellular stimulations. This process largely depends on calcium influx, calpain and cytoskeleton reorganization (Johnstone, 2006 ). Accumulating evidences suggesting that these vesicles can function as intercellular transmitters to convey their contents, in particular miRNA, from one cell to another (Rechavi et al ., 2009 ; Skog et al ., 2008 ; Valadi et al ., 2007 ). Formation and release of exosomes by cells is a complex and coordinated process which require enzymatic activation and energy (ATP) (Yáñez-Mó et al., 2015 ), and miRNA profiles of exosomes derived from bio-fluids and culture media mostly differ from their parent cells. Therefore, it is logical to think that cell may possess an active selection mechanism for exosomes and their content. Exosomes can carry a wide variety of molecules including lipids, proteins, DNAs, mRNAs and miRNAs (Simpson et al., 2012 ), among them miRNAs got the research priority because of their diversified functions and implications.
In the very first study that describe the exosome mediated transfer of miRNAs, Valadi et al. reported exosomes released from human and murine bone marrow-derived mast cells contain mRNA and miRNA, which are transferrable to other human or mouse mast cells. When exosomes from mouse mast cells transferred to human mast cells, they produce new mouse protein in recipient cells, indicating that the exosomal mRNAs are functional and they can be translated after entering into another cell (Valadi et al., 2007 ). Subsequently, numerous studies were conducted to investigate the functional relevance of exosomal ECmiRNAs. Epstein–Barr virus (EBV)-infected cells released exosomes contain ECmiRNAs and peripheral blood mononuclear cells can up take these exosomes where they involved in the repression of their target genes (Pegtel et al., 2010 ). In response to Staphylococcus aureus infection in mammary gland, milk exosomes are loaded with 14 significantly differentially expressed ECmiRNAs compared to uninfected group ( Sun et al., 2015 ). In case of renal fibrosis (outcome of chronic kidney disease), administration of erythropoietin markedly increases the expression of ECmiRNA miR-144 that was delivered to the injured renal fibroblast through exosomes to suppress tissue plasminogen activator (tPA) expression (Zhou et al., 2015 ). Furthermore, in vitro results demonstrated erythropoietin stimulates cells to release exosomes containing higher amount of miR-144 which markedly suppress the expression of tPA in cultured renal fibroblast cells (Zhou et al., 2015 ).
Due to having specific expression pattern of exosome-coupled ECmiRNAs under altered physiopathological states, it is logical to think these exosome-coupled ECmiRNAs are not passively released from the cells rather selectively loaded into exosomes to serve specific functions. Although the biological function of exosomal ECmiRNA remains questionable in vivo, numerous in vitro studies have demonstrated that ECmiRNAs enwrapped with exosomes can alter gene expression in the recipient cells. For instance, exosome derived from hypoxic leukemia cells contain a subset of upregulated miRNAs including miR-210 which my enhance tube formation in endothelial cells (Tadokoro et al., 2013 ). Another evidence for functional cell–cell transfer of miRNAs through exosomes was found when nematode derived exosomes were co-cultured with mammalian cells (mice). Administration of such exosomes results transfer of nematode miRNAs which is responsible for the repression of Dusp1 gene (Buck et al., 2014 ). Exosomes derived from biological fluids have also been shown to be involved in the repression of target genes in the recipient cells. When exosomes derived from follicular fluid of competent oocytes, enriched with specific miRNAs, administrated to granulosa cells in vitor , it results significant increase of those miRNAs in the recipient cells. Subsequently higher abundance of specific miRNAs led suppression of their target genes ( Sohel et al., 2013 ).
Release of ECmiRNA Through Microvesicles
Microvesicles (MVs), also known as ectosomes and microparticles, are membrane bound vesicles that differ from exosomes based on their mechanism of release, biogenesis and biophysical properties, including size and surface markers. While exosomes are the product of endocytic recycling pathway (inward budding), MVs are directly shed from plasma membrane through outward budding and fission of membrane vesicles. Therefore, MVs contain much similar lipid contents as the plasma membrane of their parent cells and the surface protein markers of MVs are largely dependent on the membrane of their origin (Lee et al., 2012 ). Release of MVs can take place from resting cells, however, upon stimulation the rate of secretion increases dramatically. Compare to exosomes that released in a more constitutive way and produce more homogenous populations, MVs tend to constitute more heterogeneous and larger population of vesicles, ranging from 100 to 1000 nm in diameter (Aharon et al ., 2009 ; Chironi et al ., 2009 ). Many cell types are known to secrete MVs including some cancer cells (Castellana et al., 2009 ), neurons (Marzesco et al., 2005 ) and many of the hematopoietic and vascular cell types including endothelial cells, dendritic cells and B cells (Shet, 2008 ).
Like exosomes MVs can carry variety of molecules including miRNAs. Initially MVs were considered as cellular waste, however experimental evidence suggests that MVs can influence several biological pathways and functions for example cardiovascular disorders, including atherogogenesis and thrombosis (Mack et al ., 2000 ; Mause and Weber, 2010 ; Shantsila et al ., 2010 ). The presence of miRNAs in microvesicles has now been reported from different cells including mesenchymal stem cell (Callis et al., 2009 ), mast cells (Shefler et al., 2010 ), cancer cells (Jaiswal et al., 2012 ), platelets (Hunter et al., 2008 ) and endothelial cells (Skog et al., 2008 ). When miR-143-transfected human monocytic leukemia THP-1 cells were incubation in serum-free medium, it significantly released microvesicles containing chemically modified miR-143 (Akao et al., 2011 ). Furthermore, it has been shown that embryonic stem cell derived microvesicles are miRNA enriched and they can transfer a subset of miRNAs to mouse embryonic fibroblasts in culture, suggesting that gene expression of neighboring cells might be affected by exosomal miRNA that was released by embryonic stem cells (Yuan et al., 2009 ). These findings highly support that at least some ECmiRNAs are engaged in cell–cell communication via microvesicles but still thorough investigation is needed to understand the mechanisms of selective miRNA incorporation into MVs for release.
Release of ECmiRNA Through Apoptotic Bodies
Apoptosis is an essential cellular process to remove unnecessary cells from multicellular organisms. When cells undergo apoptotic process, they release phosphatidylserine (PS) — exposed vesicles as blebs, commonly known as apoptotic bodies (ABs) or apoptotic vesicles. The major difference between ABs and cell that released other vesicles is their size. ABs are largest among all extracellular vesicles and their size reported to range from 1 to 5 μm in diameter (Turiák et al., 2011 ). Budding of MVs occurs mainly during early apoptosis and in response to stress and stimuli, whereas ABs are formed in the late stages of this death process of a cell. Membrane blebbing depends on the activity of Rho-associated coiled coil kinase 1 (ROCK1). Caspase-3 (CASP3), one of the executioner enzymes of apoptosis, was shown to be involved in the activation of ROCK1 that induces a net increase in myosin light chain phosphorylation and subsequently induces membrane blebbing (Croft, 2005 ; van der Pol et al ., 2012 ). ABs are larger than MVs & exosomes and represent the compacted or condensed residues of the apoptotic cells (Belting and Wittrup, 2008 ; Beyer and Pisetsky, 2010 ). Like other extracellular vesicles, ABs can carry a variety of molecules including miRNAs, mRNAs and fragment of DNA.
Although it was proposed several years ago that ABs carry nucleic acids (Holmgren et al., 1999 ), it was only few years ago that it has been demonstrated that ABs contain miRNAs when Zernecke et al. showed endothelial cells released Abs enriched with miR-126 and their delivery to apolipoprotein E−/− mice reduced atherosclerosis via recruitment of progenitor cells (Zernecke et al., 2009 ). Furthermore, administration of ABs containing higher number of miR-126 reduces the manifestations of atherosclerosis in mice, while miR-126-deficient Abs have no such effect (Zernecke et al., 2009 ). Interestingly, when miR-126 was found to be highly enriched only in ABs, lower abundance was reported in MVs (Hergenreider et al., 2012 ). Therefore, it is logical to speculate that specific release of ECmiRNAs into ABs may be possible under certain pathophysiological condition. However, it requires further investigation to confirm that the loading of miRNAs into apoptotic bodies are either specific & selective or whether they are nonspecifically loaded into ABs in response to a certain stimulus. Nevertheless, we cannot overrule another possibility that ECmiRNAs in ABs are by-products of dying cells and loading of these ECmiRNAs is a random process due to collapse of most biological pathways, which might be the main difference from exosome. Although the detection of miRNA within ABs is of interest, there are very few studies investigating the role AB encapsulated miRNA mediated effects in neighboring cells and more studies need to determine how specific miRNA secreted, recognized for up-take via this process.
Release of ECmiRNA Through High Density Lipoprotein
In addition to membrane vesicles enwrapped release from cells, ECmiRNAs have also been found to be released in association with lipoprotein in extracellular fluids. Lipoproteins, consist of a variety of lipids and proteins, are involved in the transportation of steroids, triglycerols, cholesterol and fat-soluble vitamins to peripheral tissues from the liver via low density lipoprotein (LDL) or removal via high density lipoprotein (HDL) (Babin and Gibbons, 2009 ). Due to their higher solubility and affinity to bind water insoluble materials, lipoproteins are able to carry nucleic acids and often used as gene delivery agents (Kim et al., 2007 ).
The idea that ECmiRNAs may present in extracellular environment in association with lipoproteins, particularly with HDL, arose when Lee et al., showed that hepatic siRNA could be effectively transferred to liver by reconstituted apolipoprotein A–I (major functional protein in HDL) (Lee et al., 2009 ). Inspired by this publication Vickers et al. demonstrated for the first time that human HDL and LDL derived from blood plasma carry a considerable amount of miRNAs (Vickers et al., 2011 ). Unlike exosomes or MVs which contain both miRNA and fragments of mRNA, highly purified HDLs are only rich in small non-coding RNAs. A specific signature of miRNAs including miR-223, miR-105 and miR-106a was identified in HDL-miRNA complexes of familial hypercholesterolemia patients (Vickers et al., 2011 ). The authors further showed that delivery of these HDL bound miRNA pool functionally downregulates their corresponding mRNA targets in cultured hepatocytes (Vickers et al., 2011 ), indicating the functional relevance of these HDL coupled ECmiRNAs. In addition, it has recently been shown that transfer of HDL rich in miR-223 significantly increases the abundance of endogenous miR-223 concentration and subsequently suppresses the expression of ICAM-1 (intracellular adhesion molecule 1) in endothelial cells and induces an anti-inflammatory action (Tabet et al., 2014 ). Interestingly, miR-223 is not transcribed in endothelial cells and HDL from miR-223−/− mice has no such effects (Tabet et al., 2014 ). In another study, Niculescu et al., demonstrated that HDL derived from patients with coronary artery disease showed specific miRNA signature compared to control (Niculescu et al., 2015 ). They found that signature of miR-486 and miR-92a associated with HDL is distinct between vulnerable and stable coronary artery disease patients and suggested that, in addition to other markers, these two ECmiRNAs can be used as a biomarker for vulnerable coronary artery disease (Niculescu et al., 2015 ). Collectively, the results of these studies indicated that ECmiRNAs coupled with HDL might have biological relevance as they exhibit distinct expression pattern in relation to different pathological conditions with biomarker potential. Although HDL mediated release and transport of ECmiRNAs is a new and exciting idea, the biggest challenge in this field is to prove that these miRNAs are actively and selectively released from donor cells and spontaneously taken up by recipient cells to mediate cell to cell communication.
Release of ECmiRNA Through Protein Complex
In addition to above mentioned pathways of release of ECmiRNAs, a handful number of studies reported that ECmiRNAs could be released through binding with Argonaut protein families, particularly Argonaut 2 (Ago2). In mammalian organisms, functional mature miRNAs generally bind with one protein complex called RNA-induced silencing complex (RISC) to regulate translation of cellular mRNAs. Ago2 is one of the major components of RISC complex. In 2011, two independent research groups reported about the presence of Ago2-miRNA complexes in cell culture media. Furthermore, western blot immunoassay shows that extracellular miRNA ultrafiltrated together with the Ago2 protein, a part of RNA-induced silencing complex, not associated with microvesicles (Turchinovich et al., 2011 ). It has also been demonstrated that only 10% cell-free miRNAs were released in plasma through micro vesicles whereas potentially 90% of the miRNAs in the circulation cofractionated with ribonucleo-protein complexes (Arroyo et al., 2011 ). Size-exclusion chromatography has been used to exclude the micro vesicle contamination from protein complexes and shows that most of the miRNA co-purified with non-vesicle-associated ribonucleo-protein complexes, only few miRNA, such as miR-16 and miR-92a associated predominantly with micro vesicles (Arroyo et al., 2011 ). Although, the role of other RNA-binding protein complexes, except Ago2, is presently unclear, collectively these results indicate that Ago2-protein complexes might be involved with the delivery of miRNA from donor cell to recipient cells and facilitate cell–cell communications.
Potential Role of ECmiRNAs
Early findings that ECmiRNAs are enwrapped with exosomes or microvesicles, along with the evidences in vitro that they can be transferred from one cell to another through exchange of exosomes (Hunter et al ., 2008 ; Mitchell et al ., 2008 ; Valadi et al ., 2007 ), provoked us to hypothesize that these vesicle-enwrapped ECmiRNAs may function as intercellular communication system in the body. Although several studies showed the possibilities of vesicle mediated transfer of ECmiRNAs covering a variety of pathophysiological conditions, it is not clear yet whether these ECmiRNAs are simply a waste product or they have any biological functions. In the previous sections of this review, it has been discussed that ECmiRNAs can be existed in extracellular milieu with association of various carriers. Since different types of ECmiRNAs are probably secreted as a result of different cellular processes, thus it is important to differentiate them in order to understand their potential roles and functions.
It is clear from the definition; ABs are formed during apoptosis from dying cells. Although, Zernecke et al., showed the relevance of ABs containing miR-126 with atherosclerosis manifestation in mice (Zernecke et al., 2009 ), it is very difficult to believe the biological viability of ABs enwrapped ECmiRNAs due to less number of reports that have been published since then. In the first study that reported the presence of protein bound ECmiRNAs, Arroyo et al., demonstrated that majority of ECmiRNAs are associated with Ago2 protein not with vesicle encapsulated (Arroyo et al., 2011 ). However, in contrast, we and others showed that majority of ECmiRNAs are concentrated in exosomes (Gallo et al ., 2012 ; Noferesti et al ., 2015 ; Sohel et al ., 2013 ). Perhaps ECmiRNAs coupled with Ago2 may be release from cells during necrosis or apoptosis (Turchinovich et al., 2011 ) and can be present in extracellular environment (Turchinovich and Burwinkel, 2012 ), but it is not known whether these Ago2-ECmiRNA complexes are also released from viable cells and can be taken up by distant cells. Therefore, it leads us to speculate that ECmiRNAs incorporated with ABs or coupled with Ago2 might solely be the waste product from dying cells or they could mediate a warning signal to the organism about cellular dysfunction. However, it does not necessarily mean that AB or Ago2 coupled ECmiRNAs present in bio-fluid are always representing the cellular dysfunction or apoptosis.
On the other hand, exosomes and microvesicles are found to be released from viable cells, though it is unclear whether they are not released from dying cells. Irrespective of cells of origin (either viable or dying), in vitro and in vivo experiments have shown that they can be taken up by a variety of viable cells. Indeed, in vitro studies have shown that ECmiRNAs can be transferred from one cell to another through exosomes and in the recipient cells they can downregulate their target genes (Kogure et al ., 2011 ; Montecalvo et al ., 2012 ; Sohel et al ., 2013 ). These findings suggesting a potential role of exosomes enwrapped ECmiRNAs that can mediate intercellular communication which might have huge impact. Recent example include, in myocardial infarction model, when exosomes derived from mesenchymal stem cells were administrated to cardiac stem cells, it results an improved migration, proliferation and angiogenic potency of cardiac stem cells (Zhang et al., 2016 ). Using in vitro model, Cui et al., demonstrated that exosomes derived from mineralizing osteoblasts are able to enter bone marrow stromal cells and subsequently, promoting them towards osteoblast differentiation through enhancing the activity of β-catenin by altering the miRNA expression in recipient cells (Cui et al., 2015 ). Exosome and microvesicles derived from cigarette smock extract — induced primary human bronchial epithelial cells contain a subset of ECmiRNA, which upon transfer promotes the myofibroblast differentiation in lung fibroblasts (Fujita et al., 2015 ). Among the other examples, transfer of exosomes from stromal cells to breast cancer cells can excite antiviral signaling and alter radiation sensitivity (Boelens et al., 2014 ), adriamycine-resistant breast cancer cells released exosomes can transmit the resistance capacity to sensitive breast cancer cells by transferring specific miRNAs (Mao et al., 2015 ), glioblastoma tumor cells released microvesicles can promote tumor growth by transferring mRNA, miRNA and protein to the normal host cells or brain microvascular endothelial cells (Skog et al., 2008 ), extracellular vesicles from myeloid neoplasm reduce the hematopoietic-supportive capacity of mesenchymal stromal cells via transfer of functional ECmiRNAs including miR-7977 (Horiguchi et al., 2016 ), and alcohol exposed monocyte derived extracellular vesicles can excite naive monocytes to differentiate into M2 macrophages by transferring ECmiRNAs including miR-27a (Saha et al., 2016 ).
In addition to orthodox role as posttranscriptional gene regulator, an unconventional role for ECmiRNAs as an activator of RNA-sensing Toll-like receptor (TLR) has recently been demonstrated. ECmiRNA let-7 was shown to activate TLR7 in neurons and induce neurodegeneration (Lehmann et al., 2012 ). Increased amount of let-7b was found in the cerebrospinal fluid of Alzheimers disease. Interestingly, administration of extracellular let-7b into the cerebrospinal fluid of wild type mice resulted neurodegeneration (Lehmann et al., 2012 ). In another study Fabbri et al. showed that tumor derived exosomes contain miR-21 and miR-29a and upon transfer to immune cells they can bind and activate TLR7 and 8 which ultimately triggered a prometastatic inflammatory response that may lead to tumor growth and metastasis (Fabbri et al., 2012 ).
Although many of the above experiments show exosome and microvesicles mediated transfer of ECmiRNAs and subsequent genetic and phenotypic changes in the recipient cells, it has recently been challenged again whether the concentration of exosomal ECmiRNAs in bio-fluids is sufficient to carry a translational repression of their target genes in the recipient cells (Chevillet et al., 2014 ). Therefore, it became necessary to precisely determine if a subset of ECmiRNAs is specifically targeted to transport to another cells via exosomes or microvesicles, gene expression patterns should be altered accordingly in the recipient cells. Otherwise in this context, ECmiRNAs enwrapped with exosomes or microvesicles present in bio-fluids would consider only be a residual amount that may not have any biological relevance.
Challenges in ECmiRNA Study
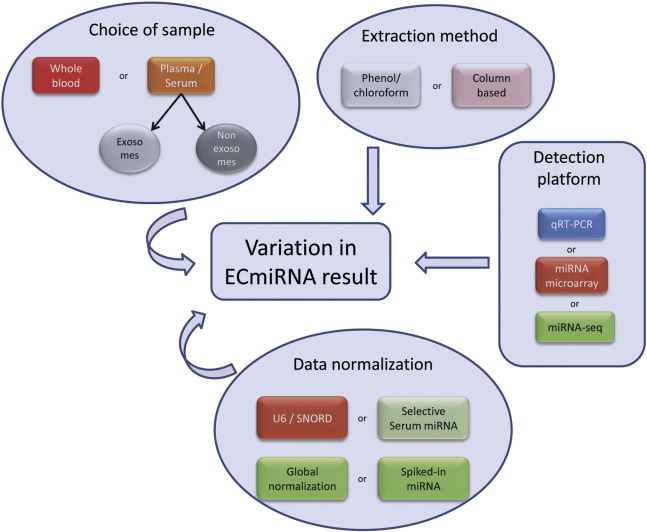
Despite the widespread interests in possible clinical application of ECmiRNAs, variations among the results have been observed and hence the use of ECmiRNAs for a reproducible and reliable biomarker development still remains in infancy. Both pre-analytical and analytical approaches, particularly choice of starting material, sample collection and processing related factors, detection platform, and finally normalization and data analysis may contribute to the variation in results. Different factors that can potentially contribute to the variations in results are shown in Fig. 3 . In order to avoid the variability among the studies, a standardize strategy should be followed from sample collection to data analysis.
|
|
|
Fig. 3. Factors contributing variation in ECmiRNA study. Several factors including starting material, extraction method, detection platform, and data analysis may directly or indirectly contribute to the variation in ECmiRNA result. |
ECmiRNAs present in circulation or other biological fluid can be quantified using different source of starting materials. For instance, when ECmiRNAs are quantified from circulation, critical decision should be made — “whole blood or serum or plasma or exosomes or microvesicles: which one is the best”? Whole blood should not be preferred, because cellular fraction (red and white blood cells) will also contribute miRNAs and influence ECmiRNA analysis (Pritchard et al., 2012 ). On the other hand, if plasma is used, EDTA will be the better choice to isolate plasma from whole blood and heparin should be avoided because it may inhibit PCR amplification. Serum can also be used as starting material for ECmiRNA study, however complete removal of cellular component is mandatory for both plasma and serum since it could impair accurate ECmiRNA quantification. Whether plasma or serum used as starting material, which fraction will be used — exosome or microvesicles or ABs or HDL or protein associated miRNAs? Exosomes are released from viable cells using a constitutive way of biogenesis and miRNA loading process (Pegtel et al., 2010 ), therefore miRNA content in exosomes may reflect a specific physio condition of an organism. Whereas, due to nonspecific release ABs, HDL & protein associated miRNAs may not be selected for biomarker study. Sample handling is another important issue. Although, ECmiRNAs are found to be stable in harsh conditions like boiling, extremely low or high pH and multiple freezing and thaw cycle, it is advisable that unnecessary freezing and thaw cycle should be avoided.
ECmiRNAs can be extracted from bio-fluids using several extraction methods. Based on the extraction method, available commercial kits can be divided in to two categories: phenol/chloroform-based extraction and column or bead based extraction method. Among the all available kits, miRNeasy Mini Kit (Qiagen) and mirVana PARIS kit (Life Technologies) produced highest yield of total RNA extraction compared to TRIzol extraction (Sourvinou et al., 2013 ). Another obstacle for ECmiRNA study is the low amount of total RNA present in bio-fluids, which makes it very difficult to precisely measure the quality and concentration of isolated total RNA. NanoDrop spectrophotometer was used by many research groups to determine the quality and quantity of ECmiRNAs extracted from bio-fluid, however if the concentration is too low (less than 50 ng/μl) NanoDrop cannot measure the concentration and sometime it remains undetected (Moret et al., 2013 ). Furthermore, in a disease state including cancer and cardiovascular disease, higher amount of miRNAs can be released to circulation than their healthy counterpart. Therefore, it is advisable to use equal volume of starting material (serum, plasma or any other biological fluid) instead of using same amount of total RNA in order to have accurate results for biomarker detection study.
Accurate quantification of ECmiRNAs from bio-fluids faced several challenges due to their low amount, short length and their GC content. Nevertheless, several detection platforms have been used to detect ECmiRNAs. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) is one of the well-established methods for detecting cellular and ECmiRNA and considered as golden standard. Among the available PCR based platform, both TaqMan TLDA microfluidic cards (Applied Biosystems) and miRCURY LNA qPCR (Exiqon) exhibit high reproducibility between technical replicates and significant correlation was observed with the two platforms (Page et al., 2013 ). Hybridization based miRNA microarrays are available from several commercial companies (Affimetrix and Agilent) and able to analyze thousands of ECmiRNAs in one assay. Compare to amplification based platform (qRT-PCR), hybridization based microarrays are less expensive; however, it typically has lower specificity and dynamic range. One of the major limitations for both qRT-PCR and Microarray is they can only detect the miRNA which are already known and annotated in miRBase. On the other hand, ECmiRNAs quantified by miRNA-seq allow us to detect both novel and known miRNAs. However, a standard protocol needs a large amount of starting material (1 μg RNA). To overcome this problem, an adopted protocol has been proposed which will provide the opportunity to obtain miRNA-seq data as little as 5 ng of total RNA extracted from bio-fluid (Williams et al., 2013 ).
After overcoming all the challenges, once we obtained data, the final challenge is to normalize and analyze the data. Housekeeping transcripts (U6 and SNORD) are generally used for the normalization of miRNA data obtained from cells or tissues, however, these housekeeping transcripts are inconsistently detected in bio-fluids due to their high RNase sensitivity. If relatively large number of miRNA is investigated, global normalization may use for data normalization. In global normalization method, a global measure of the expression profile of miRNAs, such as median or mean expression value, was used as data calibrator. However, this approach is not suitable for small number of miRNAs or for individual miRNA assay data analysis. One of the widely accepted normalization method is adding C . elegance spiked-in miRNAs during the denaturation procedure to normalize the biological variability and used for data normalization (for detail please see ( Moldovan et al ., 2014 ; Tiberio et al ., 2015 ). However, further studies are necessary to find out the systematic and best normalization method to reproducibly measure ECmiRNAs.
Concluding Remarks
Currently, we have sufficient evidences to understand the biology of ECmiRNAs and to consider ECmiRNAs, retrieved from serum/plasma or other biological fluids, as a potential regulator of developmental processes and promising biomarker for several diseases. However, inconsistent results in ECmiRNA study due to technical and experimental setup create so much controversy. It is advisable to take several important points into account before designing studies to examine ECmiRNAs. One of the important issues is low amount of miRNAs available in biofluids or cell culture media; therefore, it is important to choose the right platform where it allows analyzing ECmiRNAs with low volume of starting materials. Currently, several studies are improving the methods to retrieve the highest amount of total RNA from the fluid samples in order to profile ECmiRNAs. Another important point is the type of ECmiRNAs investigated such as exosomes, microvesicles, HDL or protein-associated miRNAs. As previously discussed, not all means of ECmiRNA release and transportation are biologically viable; therefore, it may be useful to focus on single type of ECmiRNAs, such as exosomal miRNAs or protein bound miRNAs, compared to total ECmiRNAs present in a biological fluid. Indeed, as an emerging field ECmiRNA study still require a significant technological advancement. Therefore, in order to obtain reliable results from ECmiRNA studies, development of a standardized protocol is essential. Certainly, in near future, we will be able to overcome the limitations in ECmiRNA study and it will greatly expand our understanding to developmental and disease biology and perhaps may lead to development of new therapeutics.
Compliance With Ethical Standards
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
The author declares no conflict of interest exists.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
References
- Aharon et al., 2009 A. Aharon, S. Katzenell, T. Tamari, B. Brenner; Microparticles bearing tissue factor and tissue factor pathway inhibitor in gestational vascular complications; J. Thromb. Haemost., 7 (2009), pp. 1047–1050 http://dx.doi.org/10.1111/j.1538-7836.2009.03342.x
- Akao et al., 2011 Y. Akao, A. Iio, T. Itoh, S. Noguchi, Y. Itoh, Y. Ohtsuki, T. Naoe; Microvesicle-mediated RNA molecule delivery system using monocytes/macrophages; Mol. Ther., 19 (2011), pp. 395–399 http://dx.doi.org/10.1038/mt.2010.254
- Arroyo et al., 2011 J.D. Arroyo, J.R. Chevillet, E.M. Kroh, I.K. Ruf, C.C. Pritchard, D.F. Gibson, P.S. Mitchell, C.F. Bennett, E.L. Pogosova-Agadjanyan, D.L. Stirewalt, J.F. Tait, M. Tewari; Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma; Proc. Natl. Acad. Sci. U. S. A., 108 (2011), pp. 5003–5008 http://dx.doi.org/10.1073/pnas.1019055108
- Babin and Gibbons, 2009 P.J. Babin, G.F. Gibbons; The evolution of plasma cholesterol: direct utility or a “spandrel” of hepatic lipid metabolism?; Prog. Lipid Res., 48 (2009), pp. 73–91 http://dx.doi.org/10.1016/j.plipres.2008.11.002
- Belting and Wittrup, 2008 M. Belting, A. Wittrup; Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: implications in health and disease; J. Cell Biol., 183 (2008), pp. 1187–1191 http://dx.doi.org/10.1083/jcb.200810038
- Beyer and Pisetsky, 2010 C. Beyer, D.S. Pisetsky; The role of microparticles in the pathogenesis of rheumatic diseases; Nat. Rev. Rheumatol., 6 (2010), pp. 21–29 http://dx.doi.org/10.1038/nrrheum.2009.229
- Boelens et al., 2014 M.C. Boelens, T.J. Wu, B.Y. Nabet, B. Xu, Y. Qiu, T. Yoon, D.J. Azzam, C. Twyman-Saint Victor, B.Z. Wiemann, H. Ishwaran, P.J. Ter Brugge, J. Jonkers, J. Slingerland, A.J. Minn; Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways; Cell, 159 (2014), pp. 499–513 http://dx.doi.org/10.1016/j.cell.2014.09.051
- Borchert et al., 2006 G.M. Borchert, W. Lanier, B.L. Davidson; RNA polymerase III transcribes human microRNAs; Nat. Struct. Mol. Biol., 13 (2006), pp. 1097–1101 http://dx.doi.org/10.1038/nsmb1167
- Buck et al., 2014 A.H. Buck, G. Coakley, F. Simbari, H.J. McSorley, J.F. Quintana, T. Le Bihan, S. Kumar, C. Abreu-Goodger, M. Lear, Y. Harcus, A. Ceroni, S.A. Babayan, M. Blaxter, A. Ivens, R.M. Maizels; Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity; Nat. Commun., 5 (2014), p. 5488 http://dx.doi.org/10.1038/ncomms6488
- Callis et al., 2009 T.E. Callis, K. Pandya, H.Y. Seok, R.-H. Tang, M. Tatsuguchi, Z.-P. Huang, J.-F. Chen, Z. Deng, B. Gunn, J. Shumate, M.S. Willis, C.H. Selzman, D.-Z. Wang; MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice; J. Clin. Invest., 119 (2009), pp. 2772–2786 http://dx.doi.org/10.1172/JCI36154
- Camussi et al., 2010 G. Camussi, M.C. Deregibus, S. Bruno, V. Cantaluppi, L. Biancone; Exosomes/microvesicles as a mechanism of cell-to-cell communication; Kidney Int., 78 (2010), pp. 838–848 http://dx.doi.org/10.1038/ki.2010.278
- Castellana et al., 2009 D. Castellana, C. Kunzelmann, J.-M. Freyssinet; Pathophysiologic significance of procoagulant microvesicles in cancer disease and progression; Hamostaseologie, 29 (2009), pp. 51–57
- Chen et al., 2008 X. Chen, Y. Ba, L. Ma, X. Cai, Y. Yin, K. Wang, J. Guo, Y. Zhang, J. Chen, X. Guo, Q. Li, X. Li, W. Wang, Y. Zhang, J. Wang, X. Jiang, Y. Xiang, C. Xu, P. Zheng, J. Zhang, R. Li, H. Zhang, X. Shang, T. Gong, G. Ning, J. Wang, K. Zen, J. Zhang, C.-Y. Zhang; Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases; Cell Res., 18 (2008), pp. 997–1006 http://dx.doi.org/10.1038/cr.2008.282
- Chendrimada et al., 2005 T.P. Chendrimada, R.I. Gregory, E. Kumaraswamy, J. Norman, N. Cooch, K. Nishikura, R. Shiekhattar; TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing; Nature, 436 (2005), pp. 740–744 http://dx.doi.org/10.1038/nature03868
- Cheng et al., 2014 L. Cheng, R.A. Sharples, B.J. Scicluna, A.F. Hill; Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood; J. Extracell. Vesicles, 3 (2014) http://dx.doi.org/10.3402/jev.v3.23743
- Chevillet et al., 2014 J.R. Chevillet, Q. Kang, I.K. Ruf, H.A. Briggs, L.N. Vojtech, S.M. Hughes, H.H. Cheng, J.D. Arroyo, E.K. Meredith, E.N. Gallichotte, E.L. Pogosova-Agadjanyan, C. Morrissey, D.L. Stirewalt, F. Hladik, E.Y. Yu, C.S. Higano, M. Tewari; Quantitative and stoichiometric analysis of the microRNA content of exosomes; Proc. Natl. Acad. Sci. U. S. A., 111 (2014), pp. 14888–14893 http://dx.doi.org/10.1073/pnas.1408301111
- Chim et al., 2008 S.S.C. Chim, T.K.F. Shing, E.C.W. Hung, T.-Y. Leung, T.-K. Lau, R.W.K. Chiu, Y.M.D. Lo; Detection and characterization of placental microRNAs in maternal plasma; Clin. Chem., 54 (2008), pp. 482–490 http://dx.doi.org/10.1373/clinchem.2007.097972
- Chironi et al., 2009 G.N. Chironi, C.M. Boulanger, A. Simon, F. Dignat-George, J.-M. Freyssinet, A. Tedgui; Endothelial microparticles in diseases; Cell Tissue Res., 335 (2009), pp. 143–151 http://dx.doi.org/10.1007/s00441-008-0710-9
- Croft, 2005 D.R. Croft; Actin-myosin-based contraction is responsible for apoptotic nuclear disintegration; J. Cell Biol., 168 (2005), pp. 245–255 http://dx.doi.org/10.1083/jcb.200409049
- Cui et al., 2015 Y. Cui, J. Luan, H. Li, X. Zhou, J. Han; Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression; FEBS Lett., 590 (2015), pp. 185–192 http://dx.doi.org/10.1002/1873-3468.12024
- da Silveira et al., 2012 J.C. da Silveira, D.N.R. Veeramachaneni, Q.A. Winger, E.M. Carnevale, G.J. Bouma; Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle; Biol. Reprod., 86 (2012), p. 71-71 http://dx.doi.org/10.1095/biolreprod.111.093252
- El-Hefnawy et al., 2004 T. El-Hefnawy, S. Raja, L. Kelly, W.L. Bigbee, J.M. Kirkwood, J.D. Luketich, T.E. Godfrey; Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics; Clin. Chem., 50 (2004), pp. 564–573 http://dx.doi.org/10.1373/clinchem.2003.028506
- Ender and Meister, 2010 C. Ender, G. Meister; Argonaute proteins at a glance; J. Cell Sci., 123 (2010), pp. 1819–1823 http://dx.doi.org/10.1242/jcs.055210
- Fabbri et al., 2012 M. Fabbri, A. Paone, F. Calore, R. Galli, E. Gaudio, R. Santhanam, F. Lovat, P. Fadda, C. Mao, G.J. Nuovo, N. Zanesi, M. Crawford, G.H. Ozer, D. Wernicke, H. Alder, M.A. Caligiuri, P. Nana-Sinkam, D. Perrotti, C.M. Croce; MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response; Proc. Natl. Acad. Sci. U. S. A., 109 (2012), pp. E2110–E2116 http://dx.doi.org/10.1073/pnas.1209414109
- Fujita et al., 2015 Y. Fujita, J. Araya, S. Ito, K. Kobayashi, N. Kosaka, Y. Yoshioka, T. Kadota, H. Hara, K. Kuwano, T. Ochiya; Suppression of autophagy by extracellular vesicles promotes myofibroblast differentiation in COPD pathogenesis; J. Extracell. Vesicles, 4 (2015), p. 28388
- Gallo et al., 2012 A. Gallo, M. Tandon, I. Alevizos, G.G. Illei; The majority of microRNAs detectable in serum and saliva is concentrated in exosomes; PLoS One, 7 (2012), Article e30679 http://dx.doi.org/10.1371/journal.pone.0030679
- Garzon et al., 2010 R. Garzon, G. Marcucci, C.M. Croce; Targeting microRNAs in cancer: rationale, strategies and challenges; Nat. Rev. Drug Discov., 9 (2010), pp. 775–789 http://dx.doi.org/10.1038/nrd3179
- Gilad et al., 2008 S. Gilad, E. Meiri, Y. Yogev, S. Benjamin, D. Lebanony, N. Yerushalmi, H. Benjamin, M. Kushnir, H. Cholakh, N. Melamed, Z. Bentwich, M. Hod, Y. Goren, A. Chajut; Serum microRNAs are promising novel biomarkers; PLoS One, 3 (2008), Article e3148 http://dx.doi.org/10.1371/journal.pone.0003148
- Hergenreider et al., 2012 E. Hergenreider, S. Heydt, K. Tréguer, T. Boettger, A.J.G. Horrevoets, A.M. Zeiher, M.P. Scheffer, A.S. Frangakis, X. Yin, M. Mayr, T. Braun, C. Urbich, R.A. Boon, S. Dimmeler; Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs; Nat. Cell Biol., 14 (2012), pp. 249–256 http://dx.doi.org/10.1038/ncb2441
- Holmgren et al., 1999 L. Holmgren, A. Szeles, E. Rajnavölgyi, J. Folkman, G. Klein, I. Ernberg, K.I. Falk; Horizontal transfer of DNA by the uptake of apoptotic bodies; Blood, 93 (1999), pp. 3956–3963
- Horiguchi et al., 2016 H. Horiguchi, M. Kobune, S. Kikuchi, M. Yoshida, M. Murata, K. Murase, S. Iyama, K. Takada, T. Sato, K. Ono, A. Hashimoto, A. Tatekoshi, Y. Kamihara, Y. Kawano, K. Miyanishi, N. Sawada, J. Kato; Extracellular vesicle miR-7977 is involved in hematopoietic dysfunction of mesenchymal stromal cells via poly(rC) binding protein 1 reduction in myeloid neoplasms; Haematologica (2016) http://dx.doi.org/10.3324/haematol.2015.134932
- Hossain et al., 2012 M.M. Hossain, M.M.H. Sohel, K. Schellander, D. Tesfaye; Characterization and importance of microRNAs in mammalian gonadal functions; Cell Tissue Res., 349 (2012), pp. 679–690 http://dx.doi.org/10.1007/s00441-012-1469-6
- Hunter et al., 2008 M.P. Hunter, N. Ismail, X. Zhang, B.D. Aguda, E.J. Lee, L. Yu, T. Xiao, J. Schafer, M.-L.T. Lee, T.D. Schmittgen, S.P. Nana-Sinkam, D. Jarjoura, C.B. Marsh; Detection of microRNA expression in human peripheral blood microvesicles; PLoS One, 3 (2008), Article e3694 http://dx.doi.org/10.1371/journal.pone.0003694
- Jaiswal et al., 2012 R. Jaiswal, J. Gong, S. Sambasivam, V. Combes, J.-M. Mathys, R. Davey, G.E.R. Grau, M. Bebawy; Microparticle-associated nucleic acids mediate trait dominance in cancer; FASEB J., 26 (2012), pp. 420–429 http://dx.doi.org/10.1096/fj.11-186817
- Johnstone, 2006 R.M. Johnstone; Exosomes biological significance: a concise review; Blood Cells Mol. Dis., 36 (2006), pp. 315–321 http://dx.doi.org/10.1016/j.bcmd.2005.12.001
- Kim et al., 2007 S.I. Kim, D. Shin, T.H. Choi, J.C. Lee, G.-J. Cheon, K.-Y. Kim, M. Park, M. Kim; Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I; Mol. Ther., 15 (2007), pp. 1145–1152 http://dx.doi.org/10.1038/sj.mt.6300168
- Kogure et al., 2011 T. Kogure, W.-L. Lin, I.K. Yan, C. Braconi, T. Patel; Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth; Hepatology, 54 (2011), pp. 1237–1248 http://dx.doi.org/10.1002/hep.24504
- Landthaler et al., 2004 M. Landthaler, A. Yalcin, T. Tuschl; The human DiGeorge syndrome critical region gene 8 and Its D . melanogaster homolog are required for miRNA biogenesis ; Curr. Biol., 14 (2004), pp. 2162–2167 http://dx.doi.org/10.1016/j.cub.2004.11.001
- Lee et al., 1993 R.C. Lee, R.L. Feinbaum, V. Ambros; The C . elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 ; Cell, 75 (1993), pp. 843–854
- Lee et al., 2002 Y. Lee, K. Jeon, J.-T. Lee, S. Kim, V.N. Kim; MicroRNA maturation: stepwise processing and subcellular localization; EMBO J., 21 (2002), pp. 4663–4670
- Lee et al., 2003 Y. Lee, C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Rådmark, S. Kim, V.N. Kim; The nuclear RNase III Drosha initiates microRNA processing; Nature, 425 (2003), pp. 415–419 http://dx.doi.org/10.1038/nature01957
- Lee et al., 2004 Y. Lee, M. Kim, J. Han, K.-H. Yeom, S. Lee, S.H. Baek, V.N. Kim; MicroRNA genes are transcribed by RNA polymerase II; EMBO J., 23 (2004), pp. 4051–4060 http://dx.doi.org/10.1038/sj.emboj.7600385
- Lee et al., 2009 H. Lee, S.I. Kim, D. Shin, Y. Yoon, T.H. Choi, G.-J. Cheon, M. Kim; Hepatic siRNA delivery using recombinant human apolipoprotein A–I in mice; Biochem. Biophys. Res. Commun., 378 (2009), pp. 192–196 http://dx.doi.org/10.1016/j.bbrc.2008.11.029
- Lee et al., 2012 Y. Lee, S. El Andaloussi, M.J.A. Wood; Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy; Hum. Mol. Genet., 21 (2012), pp. R125–R134 http://dx.doi.org/10.1093/hmg/dds317
- Lehmann et al., 2012 S.M. Lehmann, C. Krüger, B. Park, K. Derkow, K. Rosenberger, J. Baumgart, T. Trimbuch, G. Eom, M. Hinz, D. Kaul, P. Habbel, R. Kälin, E. Franzoni, A. Rybak, D. Nguyen, R. Veh, O. Ninnemann, O. Peters, R. Nitsch, F.L. Heppner, D. Golenbock, E. Schott, H.L. Ploegh, F.G. Wulczyn, S. Lehnardt; An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration; Nat. Neurosci., 15 (2012), pp. 827–835 http://dx.doi.org/10.1038/nn.3113
- Mack et al., 2000 M. Mack, A. Kleinschmidt, H. Brühl, C. Klier, P.J. Nelson, J. Cihak, J. Plachý, M. Stangassinger, V. Erfle, D. Schlöndorff; Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection; Nat. Med., 6 (2000), pp. 769–775 http://dx.doi.org/10.1038/77498
- Mao et al., 2015 L. Mao, J. Li, W.-X. Chen, Y.-Q. Cai, D.-D. Yu, S.-L. Zhong, J.-H. Zhao, J.-W. Zhou, J.-H. Tang; Exosomes decrease sensitivity of breast cancer cells to adriamycin by delivering microRNAs; Tumour Biol. (2015) http://dx.doi.org/10.1007/s13277-015-4402-2
- Mar-Aguilar et al., 2013 F. Mar-Aguilar, J.A. Mendoza-Ramírez, I. Malagón-Santiago, P.K. Espino-Silva, S.K. Santuario-Facio, P. Ruiz-Flores, C. Rodríguez-Padilla, D. Reséndez-Pérez; Serum circulating microRNA profiling for identification of potential breast cancer biomarkers; Dis. Markers, 34 (2013), pp. 163–169 http://dx.doi.org/10.3233/DMA-120957
- Marzesco et al., 2005 A.-M. Marzesco, P. Janich, M. Wilsch-Bräuninger, V. Dubreuil, K. Langenfeld, D. Corbeil, W.B. Huttner; Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells; J. Cell Sci., 118 (2005), pp. 2849–2858 http://dx.doi.org/10.1242/jcs.02439
- Mathivanan et al., 2010 S. Mathivanan, H. Ji, R.J. Simpson; Exosomes: extracellular organelles important in intercellular communication; J. Proteome, 73 (2010), pp. 1907–1920 http://dx.doi.org/10.1016/j.jprot.2010.06.006
- Mause and Weber, 2010 S.F. Mause, C. Weber; Microparticles: protagonists of a novel communication network for intercellular information exchange; Circ. Res., 107 (2010), pp. 1047–1057 http://dx.doi.org/10.1161/CIRCRESAHA.110.226456
- Mitchell et al., 2008 P.S. Mitchell, R.K. Parkin, E.M. Kroh, B.R. Fritz, S.K. Wyman, E.L. Pogosova-Agadjanyan, A. Peterson, J. Noteboom, K.C. O'Briant, A. Allen, D.W. Lin, N. Urban, C.W. Drescher, B.S. Knudsen, D.L. Stirewalt, R. Gentleman, R.L. Vessella, P.S. Nelson, D.B. Martin, M. Tewari; Circulating microRNAs as stable blood-based markers for cancer detection; Proc. Natl. Acad. Sci. U. S. A., 105 (2008), pp. 10513–10518 http://dx.doi.org/10.1073/pnas.0804549105
- Moldovan et al., 2014 L. Moldovan, K.E. Batte, J. Trgovcich, J. Wisler, C.B. Marsh, M. Piper; Methodological challenges in utilizing miRNAs as circulating biomarkers; J. Cell. Mol. Med., 18 (2014), pp. 371–390 http://dx.doi.org/10.1111/jcmm.12236
- Montecalvo et al., 2012 A. Montecalvo, A.T. Larregina, W.J. Shufesky, D.B. Stolz, M.L.G. Sullivan, J.M. Karlsson, C.J. Baty, G.A. Gibson, G. Erdos, Z. Wang, J. Milosevic, O.A. Tkacheva, S.J. Divito, R. Jordan, J. Lyons-Weiler, S.C. Watkins, A.E. Morelli; Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes; Blood, 119 (2012), pp. 756–766 http://dx.doi.org/10.1182/blood-2011-02-338004
- Moret et al., 2013 I. Moret, D. Sánchez-Izquierdo, M. Iborra, L. Tortosa, A. Navarro-Puche, P. Nos, J. Cervera, B. Beltrán; Assessing an improved protocol for plasma microRNA extraction; PLoS One, 8 (2013), Article e82753 http://dx.doi.org/10.1371/journal.pone.0082753
- Niculescu et al., 2015 L.S. Niculescu, N. Simionescu, G.M. Sanda, M.G. Carnuta, C.S. Stancu, A.C. Popescu, M.R. Popescu, A. Vlad, D.R. Dimulescu, M. Simionescu, A.V. Sima; MiR-486 and miR-92a identified in circulating HDL discriminate between stable and vulnerable coronary artery disease patients; PLoS One, 10 (2015), Article e0140958 http://dx.doi.org/10.1371/journal.pone.0140958
- Noferesti et al., 2015 S.S. Noferesti, M.M.H. Sohel, M. Hoelker, D. Salilew-Wondim, E. Tholen, C. Looft, F. Rings, C. Neuhoff, K. Schellander, D. Tesfaye; Controlled ovarian hyperstimulation induced changes in the expression of circulatory miRNA in bovine follicular fluid and blood plasma; J. Ovarian Res., 8 (2015), p. 81 http://dx.doi.org/10.1186/s13048-015-0208-5
- Okamura et al., 2004 K. Okamura, A. Ishizuka, H. Siomi, M.C. Siomi; Distinct roles for argonaute proteins in small RNA-directed RNA cleavage pathways; Genes Dev., 18 (2004), pp. 1655–1666 http://dx.doi.org/10.1101/gad.1210204
- Page et al., 2013 K. Page, D.S. Guttery, N. Zahra, L. Primrose, S.R. Elshaw, J.H. Pringle, K. Blighe, S.D. Marchese, A. Hills, L. Woodley, J. Stebbing, R.C. Coombes, J.A. Shaw; Influence of plasma processing on recovery and analysis of circulating nucleic acids; PLoS One, 8 (2013), Article e77963 http://dx.doi.org/10.1371/journal.pone.0077963
- Pegtel et al., 2010 D.M. Pegtel, K. Cosmopoulos, D.A. Thorley-Lawson, M.A.J. van Eijndhoven, E.S. Hopmans, J.L. Lindenberg, T.D. de Gruijl, T. Würdinger, J.M. Middeldorp; Functional delivery of viral miRNAs via exosomes; Proc. Natl. Acad. Sci. U. S. A., 107 (2010), pp. 6328–6333 http://dx.doi.org/10.1073/pnas.0914843107
- Pritchard et al., 2012 C.C. Pritchard, E. Kroh, B. Wood, J.D. Arroyo, K.J. Dougherty, M.M. Miyaji, J.F. Tait, M. Tewari; Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies; Cancer Prev. Res. (Phila.), 5 (2012), pp. 492–497 http://dx.doi.org/10.1158/1940-6207.CAPR-11-0370
- Rechavi et al., 2009 O. Rechavi, Y. Erlich, H. Amram, L. Flomenblit, F.V. Karginov, I. Goldstein, G.J. Hannon, Y. Kloog; Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small RNAs; Genes Dev., 23 (2009), pp. 1971–1979 http://dx.doi.org/10.1101/gad.1789609
- Saha et al., 2016 B. Saha, F. Momen-Heravi, K. Kodys, G. Szabo; MicroRNA cargo of extracellular vesicles from alcohol-exposed monocytes signals naive monocytes to differentiate into M2 macrophages; J. Biol. Chem., 291 (2016), pp. 149–159 http://dx.doi.org/10.1074/jbc.M115.694133
- Sang et al., 2013 Q. Sang, Z. Yao, H. Wang, R. Feng, H. Wang, X. Zhao, Q. Xing, L. Jin, L. He, L. Wu, L. Wang; Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo; J. Clin. Endocrinol. Metab., 98 (2013), pp. 3068–3079 http://dx.doi.org/10.1210/jc.2013-1715
- Shantsila et al., 2010 E. Shantsila, P.W. Kamphuisen, G.Y.H. Lip; Circulating microparticles in cardiovascular disease: implications for atherogenesis and atherothrombosis; J. Thromb. Haemost., 8 (2010), pp. 2358–2368 http://dx.doi.org/10.1111/j.1538-7836.2010.04007.x
- Shefler et al., 2010 I. Shefler, P. Salamon, T. Reshef, A. Mor, Y.A. Mekori; T cell-induced mast cell activation: a role for microparticles released from activated T cells; J. Immunol., 185 (2010), pp. 4206–4212 http://dx.doi.org/10.4049/jimmunol.1000409
- Shet, 2008 A.S. Shet; Characterizing blood microparticles: technical aspects and challenges; Vasc. Health Risk Manag., 4 (2008), pp. 769–774
- Simpson et al., 2012 R.J. Simpson, H. Kalra, S. Mathivanan; ExoCarta as a resource for exosomal research; J. Extracell. Vesicles, 1 (2012) http://dx.doi.org/10.3402/jev.v1i0.18374
- Sisco, 2001 K.L. Sisco; Is RNA in serum bound to nucleoprotein complexes?; Clin. Chem., 47 (2001), pp. 1744–1745
- Skog et al., 2008 J. Skog, T. Würdinger, S. van Rijn, D.H. Meijer, L. Gainche, M. Sena-Esteves, W.T. Curry, B.S. Carter, A.M. Krichevsky, X.O. Breakefield; Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers; Nat. Cell Biol., 10 (2008), pp. 1470–1476 http://dx.doi.org/10.1038/ncb1800
- Sohel et al., 2013 M.M.H. Sohel, M. Hoelker, S.S. Noferesti, D. Salilew-Wondim, E. Tholen, C. Looft, F. Rings, M.J. Uddin, T.E. Spencer, K. Schellander, D. Tesfaye; Exosomal and non-exosomal transport of extra-cellular microRNAs in follicular fluid: implications for bovine oocyte developmental competence; PLoS One, 8 (2013)
- Sourvinou et al., 2013 I.S. Sourvinou, A. Markou, E.S. Lianidou; Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability; J. Mol. Diagn., 15 (2013), pp. 827–834 http://dx.doi.org/10.1016/j.jmoldx.2013.07.005
- Sun et al., 2015 J. Sun, K. Aswath, S.G. Schroeder, J.D. Lippolis, T.A. Reinhardt, T.S. Sonstegard; MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection ; BMC Genomics, 16 (2015), p. 806 http://dx.doi.org/10.1186/s12864-015-2044-9
- Tabet et al., 2014 F. Tabet, K.C. Vickers, L.F. Cuesta Torres, C.B. Wiese, B.M. Shoucri, G. Lambert, C. Catherinet, L. Prado-Lourenco, M.G. Levin, S. Thacker, P. Sethupathy, P.J. Barter, A.T. Remaley, K.-A. Rye; HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells; Nat. Commun., 5 (2014), p. 3292 http://dx.doi.org/10.1038/ncomms4292
- Tadokoro et al., 2013 H. Tadokoro, T. Umezu, K. Ohyashiki, T. Hirano, J.H. Ohyashiki; Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells; J. Biol. Chem., 288 (2013), pp. 34343–34351 http://dx.doi.org/10.1074/jbc.M113.480822
- Taylor and Gercel-Taylor, 2008 D.D. Taylor, C. Gercel-Taylor; MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer; Gynecol. Oncol., 110 (2008), pp. 13–21 http://dx.doi.org/10.1016/j.ygyno.2008.04.033
- Tiberio et al., 2015 P. Tiberio, M. Callari, V. Angeloni, M.G. Daidone, V. Appierto; Challenges in using circulating miRNAs as cancer biomarkers; Biomed. Res. Int., 2015 (2015), p. 731479 http://dx.doi.org/10.1155/2015/731479
- Turchinovich and Burwinkel, 2012 A. Turchinovich, B. Burwinkel; Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma; RNA Biol., 9 (2012), pp. 1066–1075 http://dx.doi.org/10.4161/rna.21083
- Turchinovich et al., 2011 A. Turchinovich, L. Weiz, A. Langheinz, B. Burwinkel; Characterization of extracellular circulating microRNA; Nucleic Acids Res., 39 (2011), pp. 7223–7233 http://dx.doi.org/10.1093/nar/gkr254
- Turchinovich et al., 2012 A. Turchinovich, L. Weiz, B. Burwinkel; Extracellular miRNAs: the mystery of their origin and function; Trends Biochem. Sci., 37 (2012), pp. 460–465 http://dx.doi.org/10.1016/j.tibs.2012.08.003
- Turiák et al., 2011 L. Turiák, P. Misják, T.G. Szabó, B. Aradi, K. Pálóczi, O. Ozohanics, L. Drahos, A. Kittel, A. Falus, E.I. Buzás, K. Vékey; Proteomic characterization of thymocyte-derived microvesicles and apoptotic bodies in BALB/c mice; J. Proteome, 74 (2011), pp. 2025–2033 http://dx.doi.org/10.1016/j.jprot.2011.05.023
- Urbé et al., 2003 S. Urbé, M. Sachse, P.E. Row, C. Preisinger, F.A. Barr, G. Strous, J. Klumperman, M.J. Clague; The UIM domain of Hrs couples receptor sorting to vesicle formation; J. Cell Sci., 116 (2003), pp. 4169–4179 http://dx.doi.org/10.1242/jcs.00723
- Valadi et al., 2007 H. Valadi, K. Ekström, A. Bossios, M. Sjöstrand, J.J. Lee, J.O. Lötvall; Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells; Nat. Cell Biol., 9 (2007), pp. 654–659 http://dx.doi.org/10.1038/ncb1596
- van der Pol et al., 2012 E. van der Pol, A.N. Böing, P. Harrison, A. Sturk, R. Nieuwland; Classification, functions, and clinical relevance of extracellular vesicles; Pharmacol. Rev., 64 (2012), pp. 676–705 http://dx.doi.org/10.1124/pr.112.005983
- Vickers et al., 2011 K.C. Vickers, B.T. Palmisano, B.M. Shoucri, R.D. Shamburek, A.T. Remaley; MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins; Nat. Cell Biol., 13 (2011), pp. 423–433 http://dx.doi.org/10.1038/ncb2210
- Williams et al., 2013 Z. Williams, I.Z. Ben-Dov, R. Elias, A. Mihailovic, M. Brown, Z. Rosenwaks, T. Tuschl; Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations; Proc. Natl. Acad. Sci. U. S. A., 110 (2013), pp. 4255–4260 http://dx.doi.org/10.1073/pnas.1214046110
- Yáñez-Mó et al., 2015 M. Yáñez-Mó, P.R.-M. Siljander, Z. Andreu, A.B. Zavec, F.E. Borràs, E.I. Buzas, K. Buzas, E. Casal, F. Cappello, J. Carvalho, E. Colás, A. Cordeiro-da Silva, S. Fais, J.M. Falcon-Perez, I.M. Ghobrial, B. Giebel, M. Gimona, M. Graner, I. Gursel, M. Gursel, N.H.H. Heegaard, A. Hendrix, P. Kierulf, K. Kokubun, M. Kosanovic, V. Kralj-Iglic, E.-M. Krämer-Albers, S. Laitinen, C. Lässer, T. Lener, E. Ligeti, A. Linē, G. Lipps, A. Llorente, J. Lötvall, M. Manček-Keber, A. Marcilla, M. Mittelbrunn, I. Nazarenko, E.N.M. Nolte-’t Hoen, T.A. Nyman, L. O'Driscoll, M. Olivan, C. Oliveira, É. Pállinger, H.A. Del Portillo, J. Reventós, M. Rigau, E. Rohde, M. Sammar, F. Sánchez-Madrid, N. Santarém, K. Schallmoser, M.S. Ostenfeld, W. Stoorvogel, R. Stukelj, S.G. Van der Grein, M.H. Vasconcelos, M.H.M. Wauben, O. De Wever; Biological properties of extracellular vesicles and their physiological functions; J. Extracell. Vesicles, 4 (2015), p. 27066
- Yi et al., 2003 R. Yi, Y. Qin, I.G. Macara, B.R. Cullen; Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs; Genes Dev., 17 (2003), pp. 3011–3016 http://dx.doi.org/10.1101/gad.1158803
- Yuan et al., 2009 A. Yuan, E.L. Farber, A.L. Rapoport, D. Tejada, R. Deniskin, N.B. Akhmedov, D.B. Farber; Transfer of microRNAs by embryonic stem cell microvesicles; PLoS One, 4 (2009), Article e4722 http://dx.doi.org/10.1371/journal.pone.0004722
- Zernecke et al., 2009 A. Zernecke, K. Bidzhekov, H. Noels, E. Shagdarsuren, L. Gan, B. Denecke, M. Hristov, T. Köppel, M.N. Jahantigh, E. Lutgens, S. Wang, E.N. Olson, A. Schober, C. Weber; Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection; Sci. Signal., 2 (2009), p. ra81 http://dx.doi.org/10.1126/scisignal.2000610
- Zhang et al., 2002 H. Zhang, F.A. Kolb, V. Brondani, E. Billy, W. Filipowicz; Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP; EMBO J., 21 (2002), pp. 5875–5885
- Zhang et al., 2016 Z. Zhang, J. Yang, W. Yan, Y. Li, Z. Shen, T. Asahara; Pretreatment of cardiac stem cells with exosomes derived from mesenchymal stem cells enhances myocardial repair; J. Am. Heart Assoc., 5 (2016) http://dx.doi.org/10.1161/JAHA.115.002856
- Zhou et al., 2015 Zhou, Y., Fang, L., Yu, Y., Niu, J., Jiang, L., Cao, H., Sun, Q., Zen, K., Dai, C., Yang, J., 2015. Erythropoietin protects tubular basement membrane by promoting bone marrow to release extracellular vesicles containing tPA-targeting miR-144. Am. J. Physiol. Ren. Physiol. 310, ajprenal.00303.2015. doi:10.1152/ajprenal.00303.2015 .
Document information
Published on 27/03/17
Licence: Other
Share this document
Keywords
claim authorship
Are you one of the authors of this document?