Highlights
- Buprenorphine is used for opioid use disorder (OUD) management and is metabolized by the cytochrome P450 3A4 (CYP3A4) enzyme
- Pharmacogenomics studies the effect of inherited genetic variation on drug response and can personalize buprenorphine dosing
- Patients carrying the CYP3A4*1B allele may require higher than recommended doses of buprenorphine to optimize OUD management
- Key stakeholders must be consulted when establishing pharmacogenetic testing as the standard of care in OUD management
Abstract
Introduction
Opioid use disorder (OUD) is characterized by a problematic pattern of opioid use leading to clinically-significant impairment or distress. Opioid agonist treatment is an integral component of OUD management, and buprenorphine is often utilized in OUD management due to strong clinical evidence for efficacy. However, interindividual genetic differences in buprenorphine metabolism may result in variable treatment response, leaving some patients undertreated and at increased risk for relapse. Clinical pharmacogenomics studies the effect that inherited genetic variations have on drug response. Our objective is to demonstrate the impact of pharmacogenetic testing on OUD management outcomes.
Methods
We analyzed a patient who reported discomfort at daily buprenorphine dose of 24 mg, which was a mandated daily maximum by the pharmacy benefits manager. Regular urine screenings were conducted to detect the presence of unauthorized substances, and pharmacogenetic testing was used to determine the appropriate dose of buprenorphine for OUD management.
Results
At the 24 mg buprenorphine daily dose, the patient had multiple relapses with unauthorized substances. Pharmacogenetic testing revealed that the patient exhibited a cytochrome P450 3A4 ultrarapid metabolizer phenotype, which necessitated a higher than recommended daily dose of buprenorphine (32 mg) for adequate OUD management. The patient exhibited a reduction in the number of relapses on the pharmacogenetic-based dose recommendation compared to standard dosing.
Conclusion
Pharmacogenomic testing as clinical decision support helped to individualize OUD management. Collaboration by key stakeholders is essential to establishing pharmacogenetic testing as standard of care in OUD management.
Abbreviations
OUD, opioid use disorder;SUD, substance use disorder;OAT, opioid agonist treatment;PBM, pharmacy benefits manager;DSM-V, Diagnostic and Statistical Manual of Mental Disorders, 5th edition;NSDUH, National Survey on Drug Use and Health;CDC, Centers for Disease Control and Prevention;ASIPP, American Society of Interventional Pain Physicians;APA, American Psychiatric Association;ASAM, American Society of Addiction Medicine;WHO, World Health Organization;CYP3A4, cytochrome P450 3A4;PK, pharmacokinetics;PD, pharmacodynamics;PM, poor metabolizer;IM, intermediate metabolizer;EM, extensive metabolizer;UM, ultrarapid metabolizer;CLIA, Clinical Laboratory Improvement Amendments;PHM, Population Health Management
Keywords
Opioid use disorder;Opioid agonist treatment;Buprenorphine;Pharmacogenomics;Policy
1. Introduction
1.1. Epidemiology of opioid use disorders
Opioid use disorder (OUD) is defined by the Diagnostic and Statistical Manual of Mental Disorders, 5th edition (DSM-V) as “a problematic pattern of opioid use leading to clinically-significant impairment or distress” characterized by the presence of at least two criteria, such as opioid cravings, tolerance, or withdrawal, over a 12-month period (APA, 2016). OUD constitutes a significant public health crisis that affects 26.4 to 36 million people worldwide (NIDA, 2014). An estimated 2.1 million people in the United States suffered from substance use disorders (SUDs) related to prescription opioids in 2012, and an estimated 467,000 suffered from heroin dependency. The 2014 National Survey on Drug Use and Health (NSDUH) revealed a worsening epidemic, with 4.3 million Americans engaged in non-medical use of prescription painkillers in the last month while 1.4 million people used prescription painkillers non-medically for the first time in the past year (SAMHSA, 2016a). OUD increases the risk of early death, primarily from an accidental overdose, trauma, suicide, or an infectious disease, such as HIV or hepatitis C, by a factor of 20; legal problems associated with criminality and high impulsivity are also prevalent (Schuckit & Longo, 2016). Increasing nonmedical use of prescription opioids has led to a quadrupling of the number of unintentional overdose deaths in the United States since 1999 (NIDA, 2014). According to data from the Centers for Disease Control and Prevention (CDC), there are at least 44 deaths due to nonmedical use of prescription opioid pain relievers daily (SAMHSA, 2016a). There is growing evidence to suggest a relationship between increased non-medical use of opioid analgesics and heroin use in the United States (NIDA, 2014).
1.2. Management of opioid dependence
Several organizations have developed guidelines for the treatment of OUD, including the American Society of Interventional Pain Physicians (ASIPP), the American Psychiatric Association (APA), the American Society of Addiction Medicine (ASAM), and the World Health Organization (WHO). These organizations recommend a combination of pharmacological measures such as opioid agonist treatment (OAT) and psychosocial approaches such as recovery support groups to reduce illicit opioid use and harm related to opioid use while improving quality of life (WHO, 2011). According to the latest survey of opioid treatment providers, more than 300,000 people received some form of OAT for OUD in 2011 (SAMHSA, 2016b). The use of OAT in OUD management is achieved through the administration of methadone, buprenorphine, or extended-release injectable naltrexone by accredited medical professionals (WHO, 2011). These medications exert their action by occupying opioid receptors which alleviates withdrawal symptoms without inducing substantial intoxication. Methadone and buprenorphine are medications with strong clinical evidence for use in OAT because they are sufficiently long-acting for once-daily dosing, can be used in opioid withdrawal, and do not produce the cycles of intoxication and withdrawal seen in shorter-acting opioids, such as heroin (WHO, 2011).
Buprenorphine/naloxone is an opioid partial agonist/opioid antagonist combination medication indicated in the treatment of opioid dependence. Naloxone is included to discourage parenteral use. The dosing range as published in the package insert for buprenorphine/naloxone is from 4 mg/1 mg to 24 mg/6 mg, with a recommended dose of 16 mg/4 mg; doses exceeding 24 mg/6 mg are not considered to garner an additional clinical benefit (Indivior, 2015). Buprenorphine is metabolized by the cytochrome P450 3A4 (CYP3A4) enzyme. CYP3A4 is encoded by a polymorphic gene and is responsible for the metabolism of approximately 50–60% of currently prescribed medications (Rendic and Di Carlo, 1997 ; Westlind-Johnsson et al., 2006). Variation in CYP3A4 correlates to differences in metabolism rates of medications like buprenorphine. Genetic differences in genes involved in drug metabolism and response can complicate the OAT process. Novel approaches to treatment selection and dosing are warranted to overcome the challenges presented by these genetic differences. Current pharmacogenomic testing strategies can accurately identify clinically actionable variants in all related genes is one solution for optimizing drug selection and dosing for each patient.
1.3. Pharmacogenomics: individualizing OAT dosing in OUD management
Clinical pharmacogenomics is the study of effects of inherited genetic variation on an individuals medication response and combines pharmacology (the science of drug kinetics and dynamics of response) and genomics (the study of the entire genome) to optimize medication therapy (NIH, 2016a). Fig. 1 shows examples of the types of genetic mutations that may give rise to functional phenotypes. Polymorphisms in pharmacodynamic (PD) genes can affect drug action at its target, such as a receptor, and polymorphisms in pharmacokinetic (PK) genes, such as the cytochrome P450 (CYP450) family of metabolic enzymes, can affect blood and tissue drug levels. Functional variants in the CYP3A4 gene impact the rate at which drugs are metabolized and correlate to four basic phenotypes: poor metabolizers (PMs), intermediate metabolizers (IMs), extensive metabolizers (EMs), and ultra-rapid metabolizers (UMs) (Fig. 2). These pharmacogenetic variants have direct clinical application in the OAT of OUD, as buprenorphine is metabolized primarily by CYP3A4. The patients CYP3A4 metabolizer phenotype can impact treatment outcomes with buprenorphine. Patients that are CYP3A4 PMs may have higher than normal plasma levels of buprenorphine, putting the patient at risk for untoward side effects. Conversely, patients that are CYP3A4 UMs may have lower than normal serum levels of buprenorphine, which may manifest as opioid cravings and/or withdrawal symptoms. Pharmacogenetic testing conducted at the onset of treatment can guide medical practitioners, a priori, to the optimal dose for the patient by determining their metabolizer phenotype. Table 1 below explains common terminology used when describing pharmacogenomics testing results.
|
|
|
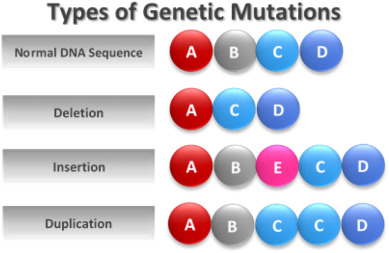
Fig. 1. Types of genetic mutations. This figure depicts a comparison of a normal DNA sequence compared to DNA sequences that became mutated during DNA replication. Each circle represents a DNA nucleotide. |
|
|
|
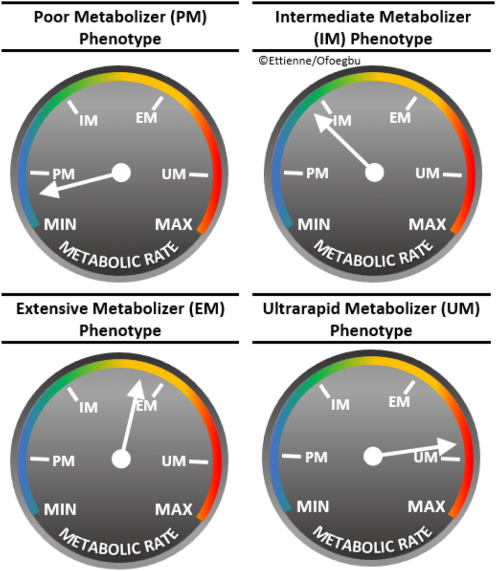
Fig. 2. CYP450 metabolizer phenotypes. These “speedometers” depict the relative differences in rate between the four metabolizer phenotype: poor metabolizer (PM), intermediate metabolizer (IM), extensive metabolizer (EM), and ultrarapid metabolizer (UM). |
| Term | Definition |
|---|---|
| Allele | Any of the alternative forms of a gene that may occur at a given locus (Merriam-Webster Dictionary, 2016a) |
| Genotype | All or part of the genetic constitution of an individual or group (Merriam-Webster Dictionary, 2016b) |
| Phenotype | The observable properties of an organism that are produced by the interaction of the genotype and the environment (such as the gene that codes for the CYP3A4 enzyme) (Merriam-Webster Dictionary, 2016c) |
The successful implementation of pharmacogenomics in clinical practice is dependent on a number of different processes, including (1) a priori knowledge of functional variants and their impact on drug metabolism and therapeutic effects, (2) the ability to accurately test a patient for known functional variants involved in drug disposition and dynamics and determine metabolizer phenotype, and (3) the ability to use this information to improve the standard of care by prescribing the right drug at the right dose.
2. Materials and methods
2.1. Case report in psychosocially-assisted pharmacological treatment of OUD: preemptive clinical pharmacogenomic testing
Below is a case report of a patient with OUD receiving buprenorphine/naloxone for OAT. The recommended dose of buprenorphine ranges from 16 to 24 mg per day. Pharmacogenetic testing was utilized as a clinical decision support tool that revealed that patients exhibiting the CYP3A4*1B genotype required higher doses of buprenorphine/naloxone than recommended in order to maintain stability.
Case Description: An African-American male has been receiving buprenorphine/naloxone for OUD management from his primary care physician for the past three years. He is married, living in a stable home, and gainfully employed. He is seen by his physician every one to four weeks. At each scheduled visit, routine urine screenings were conducted to confirm adherence to buprenorphine therapy and detect the presence of illicit and unauthorized substances. A urine screening without a detectable presence of buprenorphine suggests that the patient was not adherent to their OUD treatment regimen. The last three unauthorized substance screenings were conducted via oral swab, and the absence of buprenorphine does not suggest nonadherence to the OUD treatment regimen. Additionally, assessment of buprenorphine therapy, including dose adjustment determinations and the presence of withdrawal symptoms, were conducted at each visit. DNA was collected via buccal swab and sent to a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory for pharmacogenetic analysis.
3. Results
The patient was determined to exhibit the CYP3A4*1/*1B genotype. The patient was managed on 28 mg of buprenorphine per day and was adherent. The patient also used synthetic marijuana occasionally. In November 2015, the insurance company for the patient switched pharmacy benefits managers (PBMs). The new PBM instituted a new policy requiring prior authorization and limiting buprenorphine daily doses to 24 mg.
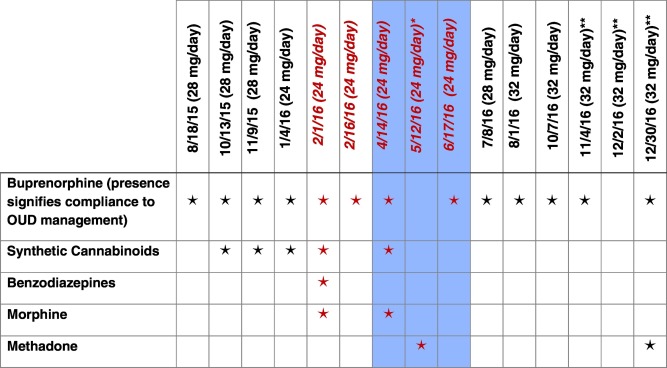
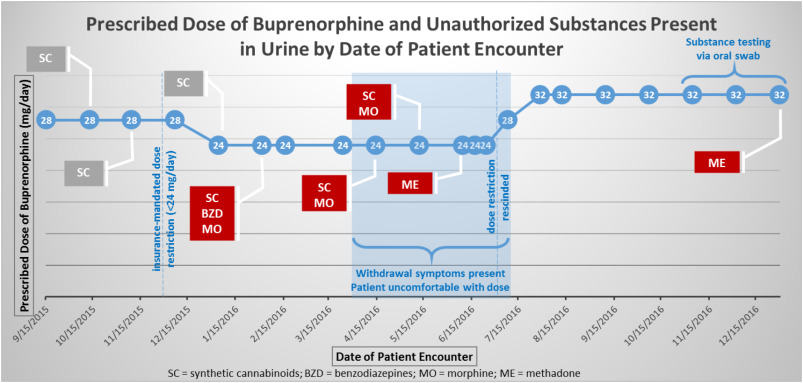
The patients daily buprenorphine dose was reduced to 24 mg from January 4, 2016 until July 8, 2016, when the insurance company rescinded the 24 mg daily dose maximum. During this period of time, the patient experienced withdrawal symptoms and reported discomfort on the lower dose. Morphine was detected in the urine four times and methadone and benzodiazepines were each detected once (Fig. 3). After the insurance-mandated dose reduction was rescinded on July 8, 2016, the patients original buprenorphine dose of 28 mg per day was restored. Due to the patient expressing discomfort, the dose was further increased to 32 mg per day. At this dose, the patient remained free of unauthorized substances until December 30, 2016, when the patient stated that he self-administered methadone acquired from a friend during the holidays. Fig. 4 illustrates the dosing changes and presence of unauthorized substances in the urine over the last year for the patient.
|
|
|
Fig. 3. Substances present in urine screen and date urine sample collected (prescribed dose of buprenorphine in mg/day). The red italicized font indicates the dose reduction to 24 mg/day of buprenorphine. Blue shading indicates period in which patient experienced withdrawal symptoms. *This urine screen indicates an absence of buprenorphine, which suggests medication nonadherence. **These substance use screenings were performed via oral swab, and the absence of buprenorphine in the results does not imply a lack of medication adherence. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) |
|
|
|
Fig. 4. Prescribed dose of buprenorphine and unauthorized substances present in urine by date of patient encounter. |
4. Discussion
There is conflicting evidence in the literature regarding the phenotype associated with the CYP3A4*1B genotype; some studies have associated the CYP3A4*1B allele with an EM phenotype, while other studies have purported that the CYP3A4*1B correlates to an UM phenotype ( Westlind-Johnsson et al., 2006). In this case report, the presence of the CYP3A4*1B allele appears to be consistent with an UM phenotype, as higher doses of buprenorphine were required for successful maintenance therapy. This case report supports existing evidence patients that have at least one copy of the CYP3A4*1B allele metabolize buprenorphine at an accelerated rate compared to the EM phenotype (CYP3A4*1/*1). This accelerated rate of metabolism may have resulted in sub-optimal therapeutic levels of buprenorphine prior to the next scheduled dose. Sub-optimal therapeutic medication levels may have subsequently resulted in the patient seeking morphine, benzodiazepines, and methadone to supplement the reduced buprenorphine dose. Once the patient was able to receive higher doses of buprenorphine, he was free of unauthorized medication use for three months by urine drug testing. It is possible that the initial dose of 28 mg of buprenorphine daily given prior to the insurance-mandated dose reduction to 24 mg daily may not have been sufficient, as evidenced by the occasional use of synthetic cannabinoids during that time; it was at the 32 mg per day buprenorphine dose that the patient was completely free of unauthorized substances.
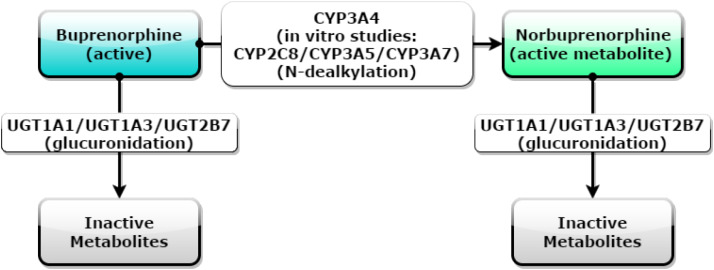
Although CYP3A4 is one of the main metabolic enzymes for buprenorphine, other proteins along the buprenorphine metabolism pathway may serve as genes of interest for pharmacogenetic testing. Fig. 5 depicts the metabolism of buprenorphine to norbuprenorphine and inactive metabolites. In addition to CYP3A4, in vitro studies have shown that the CYP2C8, CYP3A5, and CYP3A7 enzymes may contribute to buprenorphine metabolism (Picard et al., 2005). The uridine diphosphate glucuronosyl transferase (UGT) enzyme subtypes 1A1, 1A3, and 2B7 are also potential clinically-actionable targets for pharmacogenetic testing (Rouguieg et al., 2010). Furthermore, catechol-O-methyltransferase (COMT), p-glycoprotein, and the mu-opioid receptor are potential pharmacodynamic targets for pharmacogenetic testing ( Somogyi et al., 2015). Further research must be conducted to determine the effect that the aforementioned potential pharmacogenetic targets have on buprenorphine pharmacokinetics and pharmacodynamics.
|
|
|
Fig. 5. The metabolism of buprenorphine to norbuprenorphine and inactive metabolites. Buprenorphine undergoes N-dealkylation to norbuprenorphine via CYP3A4 (and CYP2C8, CYP3A5, and CYP3A7, according to in vitro studies) and glucuronidation via UGT1A1, UGT1A3, and UGT2B7 to inactive metabolites. Norbuprenorphine also undergoes glucuronidation via UGT1A1, UGT1A3, and UGT2B7 to inactive metabolites. |
4.1. Challenges in management of opioid dependence with buprenorphine/naloxone
Although buprenorphine/naloxone provides benefits to some patients, current challenges limit the potential benefit of buprenorphine/naloxone in ethnically-diverse populations. The population data used to derive the dosing recommendations for buprenorphine/naloxone is not fully generalizable to Hispanic and African-American populations. The buprenorphine/naloxone package insert cited a clinical trial in which 187 opioid-dependent men and women were assessed to provide controlled data on direct induction with buprenorphine/naloxone versus indirect buprenorphine-to-buprenorphine/naloxone induction. Although the study was used to make generalizations regarding the efficacy of the medication for the entire population, the majority of the study participants were Caucasian (95%) and male (75%) (Amass et al., 2012). In another clinical trial comparing the efficacy of buprenorphine film to sublingual tablets, the study population was comprised of 630 Caucasian participants (83.1%), 100 African-American participants (13.2%), and 28 participants classified as other or not recorded (3.7%) (Gunderson, Hjelmstrom, & Sumner, 2015). Information regarding study participant ethnicity was not included on the ClinicalTrials.gov website. This makes it more challenging to determine whether a study had adequate representation of ethnically-diverse populations. Without adequate representation of ethnically-diverse populations, it is not feasible to make generalizations regarding the efficacy of these medications in these groups.
The clinical trials used to derive the standard dosing recommendations for buprenorphine/naloxone did not take into account for possible genetic effects on buprenorphine/naloxone use. Considering that buprenorphine/naloxone is metabolized primarily via CYP3A4, the inclusion of pharmacogenomic testing during the clinical trials may have elucidated a need for variable dosing based on phenotypic presentation. The package insert makes no mention of pharmacogenomic considerations in the dosing of buprenorphine/naloxone. UMs could exhibit a sub-therapeutic clinical response and require dosing that exceeds the maximum daily recommended maintenance dose for buprenorphine/naloxone. These limitations should be addressed in order to further enable pharmacogenetic testing implementation for OUD management.
5. Conclusion
5.1. Application of pharmacogenomics informed by a population health management approach to care
Population Health Management (PHM) is the proactive application of strategies and interventions to defined groups in order to improve the health of the individuals within the population in a clinically-effective, cost-effective, and safe manner. From a PHM perspective, there are several benefits to expanding the use of pharmacogenetic testing and genomic registries. Treatment selection and dosing based on the pharmacogenetic phenotype of individual patients results in: (1) increased efficacy, (2) increased adherence, (3) improved outcomes, and (4) increased pharmacoeconomic benefit (Rendic & Di Carlo, 1997). In the case report presented, pharmacogenetic testing helped the physician determine the appropriate dose of buprenorphine/naloxone for the patient, which led to improved adherence and decreased relapse to unauthorized opioid use. The potential exists to move towards more effective treatment, outcomes, and recovery, which are measured in two spheres for SUD treatment:
- Individual outcomes
- Subjective – reduce or eliminate intense withdrawal and cravings associated with opioids
- Objective - time in OAT and recovery as measured by testing negative for unauthorized substances
- Community outcomes
- Reduced medical costs through decreased exposure to epidemic infectious diseases like hepatitis C (HCV) and human immunodeficiency virus (HIV) infection
- Decreased emergency visits, hospitalizations, and re-hospitalizations
- Family reintegration and employment
- Reduction in drug seeking-related crime and the resulting cost of policing and incarceration.
Pharmacogenetic testing implementation as a standard of care in OUD management can be realized through the collaborative efforts of key stakeholders, such as pharmacists, physicians, patients academic medical centers, regulatory agencies, funding agencies, payers, and the pharmaceutical industry (Issa, 2013). Physicians and pharmacists in academic medical centers and specially-trained community-based provider teams can collectively provide pharmacogenetic testing and result interpretation for optimal buprenorphine dosing.
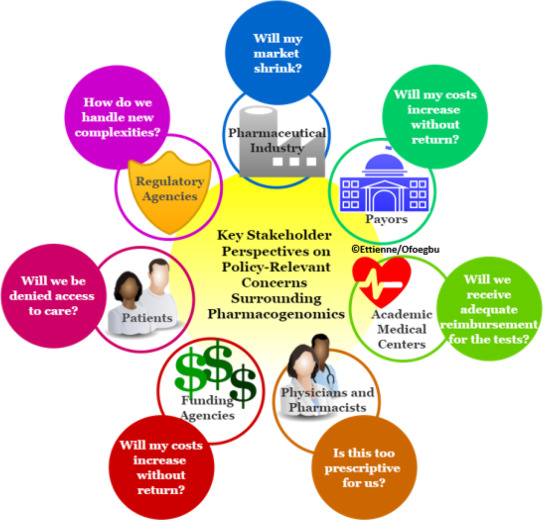
Key stakeholders may have concerns regarding the widespread adoption of pharmacogenetic testing (see Appendix A). Payers and funding agencies may be faced with costs for testing, counseling, and education; physicians may deem the testing too prescriptive; patients may raise ethical issues regarding the collection of and access to their genomic data, and government agencies may face new regulatory complexities in the oversight of health care (Issa, 2013). These concerns will present challenges in the progress of the adoption and incorporation of pharmacogenomic testing in OUD management.
5.2. Progress in clinical pharmacogenomic policy
In January 2013, the Food and Drug Administration (FDA) issued a guidance document entitled “Clinical Pharmacogenomics: Premarket Evaluation in Early-Phase Clinical Studies and Recommendations for Labeling” (FDA, 2013). The FDA recommended the use of pharmacogenomics in early-phase clinical studies to (1) identify populations that should receive variable dosing based on drug response, (2) identify responder populations based on phenotypic, receptor, or genetic characteristics, and (3) identify groups of patients at high risk for adverse effects (FDA, 2013). The FDA also recommended that medication labeling include pharmacogenomic information such as (1) the frequency of alleles, genotypes, haplotypes, or other genomic markers of relevance, (2) a description of the functional effects of genomic variants, (3) the effect of genotype on important pharmacokinetic (PK) parameters on pharmacodynamic (PD) endpoints, (4) a description of pharmacogenomic studies that provided evidence of genetically-based differences in drug benefit or risk, and (5) dosing and patient selection recommendations based on genotype (FDA, 2013). As a result, some drug manufacturers have included pharmacogenetics in clinical trials and have included the information in medication package inserts. The FDA currently has pharmacogenomic recommendations for more than 200 medications. Of these, 36 are classified as “genetic testing required,” 6 medications are classified as “genetic testing recommended,” and 79 medications are classified as having “actionable pharmacogenomics” with recommendations on medications adjustments that affect toxicity and efficacy (Whirl-Carrillo et al., 2012).
On January 20, 2015, President Obama announced the Precision Medicine Initiative (PMI) in his State of the Union address and called for $215 million to be allocated towards extending precision medicine to all diseases. A prominent example is the building of a national, large-scale cancer genomics research participant cohort at the National Cancer Institute (NIH, 2016b). The advent of the PMI has the potential to increase the ethnic and genetic diversity of samples in pharmacogenomic testing algorithms and improve the outcomes for the treatment of OUD in ethnically-diverse populations.
5.3. Recommendations for the enhancement of clinical pharmacogenomics policy
In order to improve pharmacological treatment of OUD, further advances must be taken to improve the utilization of pharmacogenomics. Policy recommendations for the inclusion of pharmacogenomic considerations in OUD management include the following:
- Determining challenges to implementation from the perspective of each of the key stakeholders and provide resolutions to the purported challenges
- Establishing genetic polymorphisms databases with a greater proportion of underrepresented populations to improve health equity in pharmacogenomic testing algorithms
- Encouraging buprenorphine/naloxone manufacturers to provide medication strengths consistent with higher dosing in patients with ultrarapid metabolizer phenotypes for ease of administration and to provide appropriate pharmacogenomic package insert information based on FDA recommendations
- Developing clinical pathways for the use of pharmacogenomics at the bedside by using aggregated pharmacogenomic data to predict the appropriateness of testing. It is reasonable to conduct pharmacogenomic testing prior to the initiation of buprenorphine/naloxone therapy in order to determine an appropriate starting and maintenance dose. The use of clinical pathways can help with cost containment by limiting the use of pharmacogenomics testing to instances in which it is deemed medically necessary
- Redefining pharmacoeconomic analyses for opioid dependence costs by taking into account the use of pharmacogenomic testing and the savings associated with the prevention of downstream cost that can be ameliorated with customized medicine
- Developing one universal piece of legislation regarding the collective approach of the government towards the management of opioid dependence
- Including ethnicity information directly on the ClinicalTrials.gov website
Acknowledgements
The authors wish to thank Drs. Albert G. B. Amoah, Arthur C. Sackeyfio, Allen Steele Dadzie, the Howard University National Human Genome Center, the Howard University Research Centers for Minority Institutions (RCMI), and the Minority Health and Health Disparities International Research Training Program (MHIRT) for their support in our research endeavors.
Appendix A. Key stakeholder perspectives on policy-relevant concerns surrounding pharmacogenomics
Each circle set signifies a key stakeholder and a question illustrating a point of concern. These points of concern must be addressed in order to have pharmacogenomic testing as standard of care.
References
- Amass et al., 2012 L. Amass, et al.; A prospective, randomized, multicenter acceptability and safety study of direct buprenorphine/naloxone induction in heroin-dependent individuals; Addiction, 107 (1) (2012), pp. 142–151
- APA, 2016 APA; Opioid Use Disorder: Diagnostic Criteria; Retrieved from http://pcssmat.org/wp-content/uploads/2014/02/5B-DSM-5-Opioid-Use-Disorder-Diagnostic-Criteria.pdf (2016)
- FDA, 2013 FDA; CDER (Ed.), Guidance for Industry Clinical Pharmacogenomics: Premarket Evaluation in Early-phase Clinical Studies and Recommendations for Labeling (2013), pp. 1–26
- Gunderson et al., 2015 E.W. Gunderson, P. Hjelmstrom, M. Sumner; Effects of a higher-bioavailability buprenorphine/naloxone sublingual tablet versus buprenorphine/naloxone film for the treatment of opioid dependence during induction and stabilization: A multicenter, randomized trial; Clinical Therapeutics, 37 (10) (2015), pp. 2244–2255
- Indivior, 2015 Indivior; Suboxone Prescribing Information; (2015)
- Issa, 2013 A.M. Issa; Pharmacogenomics and Health Policy; (2013)
- Merriam-Webster Dictionary, 2016a Merriam-Webster Dictionary; Definition of ALLELE; Retrieved from http://www.merriam-webster.com/dictionary/allele (2016)
- Merriam-Webster Dictionary, 2016b Merriam-Webster Dictionary; Definition of GENOTYPE; Retrieved from http://www.merriam-webster.com/dictionary/genotype (2016)
- Merriam-Webster Dictionary, 2016c Merriam-Webster Dictionary; Definition of PHENOTYPE; Retrieved from http://www.merriam-webster.com/dictionary/phenotype (2016)
- NIDA, 2014 NIDA; Americas Addiction to Opioids: Heroin and Prescription Drug Abuse; Retrieved from https://www.drugabuse.gov/about-nida/legislative-activities/testimony-to-congress/2016/americas-addiction-to-opioids-heroin-prescription-drug-abuse (2014)
- NIH, 2016a NIH; What is pharmacogenomics?; Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/ (2016)
- NIH, 2016b NIH; Precision Medicine Initiative; Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/ (2016)
- Picard et al., 2005 N. Picard, et al.; In vitro metabolism study of buprenorphine: Evidence for new metabolic pathways; Drug Metabolism and Disposition, 33 (5) (2005), pp. 689–695
- Rendic and Di Carlo, 1997 S. Rendic, F.J. Di Carlo; Human cytochrome P450 enzymes: A status report summarizing their reactions, substrates, inducers, and inhibitors; Drug Metabolism Reviews, 29 (1–2) (1997), pp. 413–580
- Rouguieg et al., 2010 K. Rouguieg, et al.; Contribution of the different UDP-glucuronosyltransferase (UGT) isoforms to buprenorphine and norbuprenorphine metabolism and relationship with the main UGT polymorphisms in a bank of human liver microsomes; Drug Metabolism and Disposition, 38 (1) (2010), pp. 40–45
- SAMHSA, 2016a SAMHSA; Opioids | SAMHSA; Retrieved from http://www.samhsa.gov/atod/opioids (2016)
- SAMHSA, 2016b SAMHSA; Treatments for Substance Use Disorders; 2014-09-30T20:07-04:00; Retrieved from http://www.samhsa.gov/treatment/substance-use-disorders#opioid (2016)
- Schuckit and Longo, 2016 M.A. Schuckit, D.L. Longo; Treatment of Opioid-use Disorders; http://dx.doi.org/10.1056/NEJMra1604339 (2016)
- Somogyi et al., 2015 A.A. Somogyi, et al.; Pharmacogenetics of opioid response; Clinical Pharmacology and Therapeutics, 97 (2) (2015), pp. 125–127
- Westlind-Johnsson et al., 2006 A. Westlind-Johnsson, et al.; Identification and characterization of CYP3A4*20, a novel rare CYP3A4 allele without functional activity; Clinical Pharmacology and Therapeutics, 79 (4) (2006), pp. 339–349
- Whirl-Carrillo et al., 2012 M. Whirl-Carrillo, et al.; Pharmacogenomics knowledge for personalized medicine; Clinical Pharmacology and Therapeutics, 92 (4) (2012), pp. 414–417
- WHO, 2011 WHO; Treatment of Opioid Dependence; WHO (2011) Retrieved from http://www.who.int/substance_abuse/activities/treatment_opioid_dependence/en/
Document information
Published on 26/05/17
Submitted on 26/05/17
Licence: Other
Share this document
Keywords
claim authorship
Are you one of the authors of this document?