Summary
Background
Single nucleotide polymorphisms (SNPs) of interleukin-28B (IL28B) were associated with sustained virological response (SVR) in hepatitis C virus genotype 1 (HCV-1) infected patients treated with a standard 48-week regimen of peginterferon and ribavirin combination. Whether IL28B SNP genotype would be the influential prognosticator for patients treated with response-guided therapy (RGT) is still not well understood.
Aims
To investigate the impact of IL28B rs809917 genotype on HCV-1 infected patients treated with RGT.
Methods
A total of 128 consecutive treatment-naïve HCV-1 infected patients between July 2006 and July 2011 were analyzed. For rapid virological response (RVR) patients, we allowed an abbreviated 24-week regimen regardless of baseline viral loads; otherwise, a 48-week regimen was implemented (for patients with early virological response). The IL28B rs8099917 SNP genotypes were determined accordingly.
Results
A total of 117 patients (91.4%) were of rs8099917 TT genotype and 11 (8.6%) were of GT/GG genotype. Eighty-two of the 128 (64.1%) patients achieved SVR, occurring in 54 of 67 RVR patients (80.6%) and 28 of 61 non-RVR patients (45.9%, p < 0.001). Compared to the GT/GG genotype, patients with the TT genotype had significantly higher SVR rates (67.5% vs. 27.3%; p = 0.008) and low relapse rates (28.2% vs. 70.0%; p = 0.006). The multivariate analysis showed that RVR (odds ratio, 4.51; 95% confidence interval, 1.87–10.90; p = 0.001) and rs8099917 TT genotype (odds ratio, 6.91; 95% confidence interval, 1.53–31.17; p = 0.012) were independent factors associated with SVR.
Conclusion
For HCV-1 infected patients who were treated with RGT, the IL28B unfavorable genotype predicted a higher relapse rate; RVR and IL28B favorable genotype were independent factors associated with SVR in patients treated with RGT.
Keywords
Hepatitis C virus (HCV) ; Interleukin-28B (IL28B) ; Response-guided therapy (RGT) ; Single nucleotide polymorphisms (SNPs)
Introduction
Hepatitis C virus (HCV) infection, a leading cause of end-stage liver disease such as cirrhosis or hepatocellular carcinoma, affects approximately 170 million individuals worldwide [1] . Prevention of new HCV infection and treatment of chronic hepatitis C should be the primary goals to solve this clinical exigency. Large pivotal randomized controlled clinical trials have shown that a combination treatment with peginterferon alpha-2a or alpha-2b and ribavirin leads to a sustained virological response (SVR) rate of 50% or higher [2] ; [3] ; [4] .
The duration of treatment should be based on the HCV genotype, and 48 weeks of treatment for patients with HCV genotype 1 (HCV-1) infection was suggested as the standard of care [1] ; [4] . Rapid virological response (RVR), defined as undetectable serum HCV at Week 4 of treatment, is emerging as an important milestone in the treatment of patients with chronic hepatitis C [1] ; [5] . The integration of RVR into treatment decision may identify the patients for whom a truncated course of therapy is appropriate. By contrast, confirmation of no RVR in spite of early virological response (EVR; defined as negative or 2-log10 decline from baseline of serum HCV RNA at Week 12) allows clinicians to select the so-called slow responders among HCV genotype 1 infected patients, for whom 48–72 weeks of therapy could be beneficial [5] ; [6] ; [7] ; [8] ; [9] ; [10] ; [11] ; [12] . Such response-guided therapy (RGT), a dynamic algorithm that involves individualized treatment based on the on-treatment virological response, is widely accepted in current prevailing guidelines [1] .
Recent genome-wide associated studies have demonstrated strong evidence that single-nucleotide polymorphisms (SNPs) of interleukin-28B (IL28B) were significantly correlated with SVR when patients were treated with a standard duration therapy [13] ; [14] ; [15] ; [16] ; [17] ; [18] ; [19] ; [20] ; [21] . Nevertheless, only a few studies have focused on whether IL28B SNP phenotypes would be influential in achieving SVR in patients treated with RGT [22] . The aim of this study was to investigate the predictors of SVR in patients treated with RGT in a real-life setting, in a single nontertiary regional hospital in middle Taiwan. We particularly addressed the impact of IL28B genotype on treatment responses.
Methods
Background: real-world setting in Taiwan and our daily practice
Since October 2003, the Bureau of National Health Insurance (BNHI) in Taiwan has instituted a hepatitis C trial treatment program that reimburses the cost for a standard 24-week combination therapy of peginterferon and ribavirin regardless of genotypes. The concept of on-treatment virological monitoring was not integrated into management with standard therapy in our daily practice until July 2006. After the introduction of the RGT concept, we urged our patients to monitor their hepatitis C viral loads during treatment. For HCV-1 infected patient who achieved an RVR, we allowed an abbreviated 24-week regimen regardless of baseline viral loads; otherwise, patients received the 48-week regimen at their own expense for extra medications from the 25th week to the 48th week of treatment. The BNHI finally agreed to a 48-week regimen for patients without RVR (for details of on-treatment viral monitoring, see below) after November 2009.
Patients and therapeutic regimens
This study comprised 128 consecutive treatment-naïve HCV-1 infected patients between July 2006 and July 2011. Eligible participants were patients with positive serum antibody to HCV, who were older than 18 years, who had elevated alanine aminotransferase, and who had HCV RNA detectable in their sera by commercial nucleic acid tests. Patients were excluded if they have mixed HCV genotypes infection, concurrent infections of hepatitis B virus (positive hepatitis B surface antigen test), decompensated cirrhosis, neutropenia (<1.5 × 109 /L), thrombocytopenia (<80 × 109 /L), anemia (<12 g/dL), hepatocellular carcinoma, and uncontrolled thyroid function. Informed consent was obtained from all patients. The study was carried out in accordance with the provisions of the 1975 Declaration of Helsinki and with the approval of the Institutional Review Board of the hospital.
The starting dose of peginterferon alpha-2b was 1.5 μg/kg/wk, and this was adjusted by 0.5-μg/kg decrements in patients who developed adverse events attributable to interferon. For patients receiving peginterferon alpha-2a, the starting dose was fixed at 180 μg/wk, and this was reduced by 45 μg/wk if needed. The starting dose of ribavirin was adjusted by body weight and computed as follows: <75 kg, 1000 mg/d; and ≥75 kg, 1200 mg/d. In general, we reduced the dose of ribavirin by 200-mg decrements in patients whose hemoglobin declined to <10 g/dL or in those patients who developed other adverse effects attributable to this drug. Erythropoietin treatment was permitted if the hemoglobin level was <10 g/dL.
Laboratory methods
Serum HCV RNA levels were determined by a quantitative polymerase chain reaction assay (Cobas TaqMan HCV Test version 2.0; Roche Diagnostics GmbH, Mannheim, Germany; detection limit, 15 IU/mL). In our analysis, patients with a baseline HCV RNA level of >600,000 IU/mL were considered as having a high viral load. Human genomic DNA was extracted from peripheral blood mononuclear cells by QIAamp DNA Blood Mini Kit (Qiagen Inc., Valencia, CA, USA). For IL28B rs8099917 SNP genotype determination, the TaqMan 5′ nuclease SNP genotyping assays were used. The samples were analyzed using the Applied Biosystems StepOne and StepOnePlus Real-Time PCR Systems (Applied Biosystems Inc., Foster City, CA, USA). All assay reagents including primers and probes were purchased from Applied Biosystems Inc. The discrimination between wild-type and mutant alleles was achieved based on labeling the probes with two different dyes: VIC for wild-type and FAM for mutant alleles. Because routine fibrosis assessments obtained from liver biopsy are not required by BNHI, we used the aspartate aminotransferase (AST)/platelet ratio index (APRI) to represent the severity of liver fibrosis, which was calculated using the following equation:
|
|
Assessment of adherence and efficacy
Patients who received ≥80% of both their total interferon and ribavirin doses for ≥80% of the expected duration of therapy were considered to be 80/80/80 adherent. The treatment responses were determined using the intention-to-treat (ITT) analysis, and we included any patient who had taken at least one dose of medications in this study. RVR was defined as undetectable serum HCV at Week 4 of treatment. Complete (cEVR) and partial early virological response (pEVR) were defined as negative or 2-log10 decline from baseline of serum HCV RNA at Week 12. A delayed virological response was defined as undetectable HCV RNA at Week 24 in patients with pEVR. “Nonresponse” was defined as a drop in HCV RNA level of <2 log10 from baseline at Week 12. Lastly, the end-of-treatment virological response (EOTVR) and SVR were defined as undetectable HCV RNA at the end of treatment (EOT) and Week 24 after EOT.
Statistical analysis
The descriptive statistics used were mean ± standard deviation for continuous variables and proportion for categorical variables. The analysis was conducted using two-sample t test, Pearsons Chi-square test, and Fishers exact test when appropriate. All statistical testing was two-tailed at the 5% level. Logistic regression models were used to evaluate possible predictors of treatment responses, and results were reported as odds ratio (OR) and their 95% confidence intervals (CIs). Covariates with a two-sided p < 0.1 at univariate analysis were included in a multivariate model, to determine the independent determinants. For all tests, p < 0.05 was considered statistically significant. The analysis software used was SPSS, version 17.0 (SPSS Inc., Chicago, IL, USA).
Results
Patient profiles, stratified by IL28B rs8099917 genotypes
The IL28B rs8099917 genotypic analysis showed that 117 patients (91.4%) carried the TT genotype and the remaining 11 patients (8.6%) carried the GT/GG genotype. The basic demographic, virological, and clinical features were similar between the patients carrying the rs8099917 TT and GT/GG genotypes (Table 1 ).
| Characteristics | rs8099917 | ||
|---|---|---|---|
| Genotype TT (n = 117) | Genotype GT/GG (n = 11) | p | |
| Age, y | 57.5 ± 10.3 | 57.0 ± 8.9 | 0.898 |
| Male sex | 64 (54.7) | 6 (54.5) | 0.992 |
| BMI | 25.4 ± 3.7 | 25.5 ± 4.7 | 0.978 |
| ALT, IU/L | 118.1 ± 84.3 | 150.7 ± 90.2 | 0.224 |
| GGT, IU/L | 87.8 ± 165.4 | 126.8 ± 165.6 | 0.477 |
| APRI | 1.9 ± 1.7 | 1.8 ± 1.4 | 0.890 |
| Creatinine, mg/dL | 0.9 ± 0.3 | 0.9 ± 0.2 | 0.688 |
| DM | 19 (16.2) | 0 (0) | 0.148 |
| Baseline HCV RNA | |||
| In log IU/mL | 6.1 ± 0.9 | 6.1 ± 0.8 | 0.968 |
| <600,000 IU/mL | 37 (31.6) | 5 (45.5) | 0.350 |
Data are presented as mean ± SD or n (%).
ALT = alanine aminotransferase; APRI = aspartate aminotransferase (AST)/platelet ratio index; BMI = body mass index; DM = diabetes mellitus; GGT = gamma-glutamyl transpeptidase; SD = standard deviation.
Virological responses
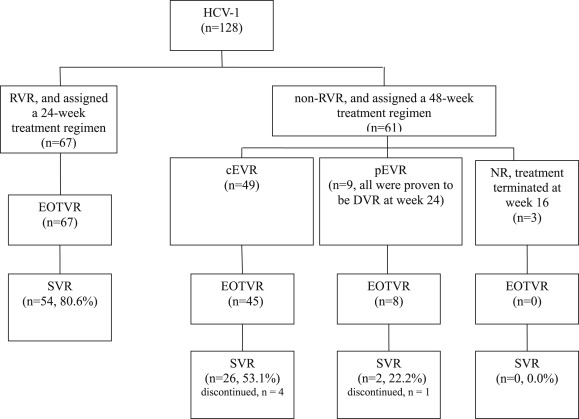
Of the 128 patients, five had premature discontinuation of treatment. Thus, the discontinuation rate for this series was 3.9%. An outline of the flow diagram used in this study is shown in Fig. 1 . Eighty-two of the 128 (64.1%) patients achieved SVR, occurring in 54 of 67 RVR patients (80.6%) and 28 of 61 non-RVR patients (45.9%, p < 0.001). For patients without RVR, 49 individuals achieved the goal of cEVR, and nine attained delayed virological response; the remaining three patients were determined to be nonresponsive, and the treatment was terminated at Week 16.
|
|
|
Figure 1. Flow diagram illustrating the treatment outcome. cEVR = complete virological response; DVR = delayed virological response; EOTVR = end-of-treatment virological response; NR = nonresponse; pEVR = partial virological response; RVR = rapid virological response; SVR = sustained virological response. |
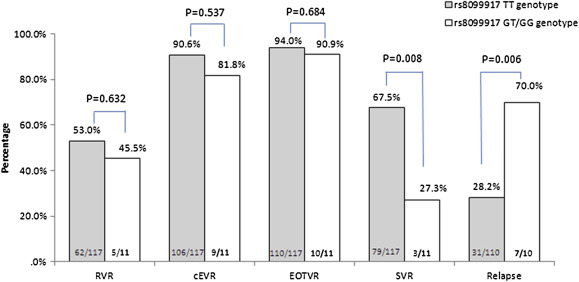
The RVR, cEVR, and EOTVR rates were not different between patients with IL28B rs8099917 TT and GT/GG genotypes (Fig. 2 ). However, compared to the G carrying allele (GT/GG), those with the homozygous TT genotype had a significantly higher SVR rates (67.5% vs. 27.3%; p = 0.008) and low relapse rates (28.2% vs. 70.0%; p = 0.006; Fig. 2 ).
|
|
|
Figure 2. Treatment response rates with different IL28B rs8099917 genotypes. The Arabic numerals at the base of the bar indicate the patient numbers. The definition of cEVR was anyone who cleared serum HCV at Week 12, regardless of the achievement of RVR. cEVR = complete virological response; EOTVR = end-of-treatment virological response; RVR = rapid virological response; SVR = sustained virological response. |
Predictors of RVR
As shown in Table 2 , the univariate analysis demonstrated that male sex, lower body mass index, and lower baseline HCV RNA level <600,000 IU/mL were significantly associated with higher RVR rates (Table 2 ). Further multivariate analysis revealed that both male sex (OR 5.25; 95% CI, 2.11–13.07; p < 0.001) and baseline HCV viral loads <600,000 IU/mL (OR 7.75; 95% CI, 2.76–21.74; p < 0.001) were predictors of an RVR. Noticeably, the IL28B rs8099917 genotype did not have any impact on the chance of RVR in this study.
| Univariate analysis | Multivariate analysisa | ||||||
|---|---|---|---|---|---|---|---|
| RVR (n = 67) | Non-RVR (n = 61) | p | OR | 95% CI | p | ||
| Male sex | 47 (70.1) | 23 (37.7) | <0.001 | 5.25 | 2.11 | 13.07 | <0.001 |
| BMI <25 | 38 (56.7) | 24 (39.) | 0.050 | 1.06 | 0.45 | 2.51 | 0.895 |
| ALT <80 IU/L | 21 (31.3) | 23 (37.3) | 0.449 | ||||
| GGT <58 IU/L | 37 (61.7) | 26 (46.4) | 0.100 | 2.18 | 0.92 | 5.16 | 0.077 |
| Baseline HCV RNA level, <600,000 IU/mL | 34 (50.7) | 8 (13.1) | <0.001 | 7.75 | 2.76 | 21.74 | <0.001 |
| Age <55 y | 26 (38.8) | 22 (36.1) | 0.749 | ||||
| IL28B rs8099917 genotype TT (vs. GT/GG) | 62 (92.5) | 55 (90.5) | 0.632 | ||||
| APRI, mean ± SD | 1.8 ± 1.7 | 2.0 ± 1.7 | 0.533 | ||||
Data are presented as n (%) unless otherwise indicated.
ALT = alanine aminotransferase; APRI = aspartate aminotransferase (AST)/platelet ratio index; BMI = body mass index; CI = confidence interval; GGT = gamma-glutamyl transpeptidase; OR = odds ratio; RVR = rapid virological response; SD = standard deviation.
a. Only covariates with p < 0.1 at univariate analysis were included in a multivariate model to determine independent determinants.
Predictors of SVR
As shown in Table 3 , patients with IL28B rs8099917 TT genotype, HCV RNA level <600,000 IU/mL at baseline, and the achievement of an RVR were factors predictive of SVR in the univariate analysis. We then conducted the multivariate analyses either by baseline factors only (Model 1, see Table 3 ) or by incorporating the achievement of RVR into to the baseline factors (Model 2, see Table 3 ). In Model 1, the result showed that baseline viral load <600,000 IU/mL (OR 2.77; 95% CI, 1.14–6.75; p = 0.025) and the carriage of IL28B rs8099917 TT genotype (OR 7.06; 95% CI, 1.66–30.10; p = 0.008) were independent factors associated with SVR. In Model 2, only the attainment of an RVR (OR 4.51; 95% CI, 1.87–10.90; p = 0.001) and the carriage of rs8099917 TT genotype (OR 6.91; 95% CI, 1.53–31.17; p = 0.012) were independent factors associated with SVR, but not low viral load ( Table 3 ).
| Univariate analysis | Multivariate analysis, Model 1 | Multivariate analysis, Model 2a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SVR (n = 82) | Non-SVR (n = 46) | p | OR | 95% CI | p | OR | 95% CI | p | |||
| Male sex | 49 (59.8) | 21 (45.7) | 0.124 | ||||||||
| BMI <25 | 41 (50.0) | 21 (45.7) | 0.637 | ||||||||
| ALT <80 IU/L | 26 (31.7) | 18 (39.1) | 0.396 | ||||||||
| GGT <58 IU/L | 39 (51.3) | 24 (60.0) | 0.372 | ||||||||
| Baseline HCV RNA level, <600,000 IU/mL | 32 (39.0) | 10 (21.7) | 0.046 | 2.77 | 1.14 | 6.75 | 0.025 | 1.49 | 0.55 | 4.04 | 0.437 |
| Age <55 y | 33 (40.2) | 15 (32.6) | 0.392 | ||||||||
| Adherence (80/80/80 rule) | 67 (81.7) | 32 (69.6) | 0.115 | ||||||||
| With RVR | 54 (65.9) | 13 (28.3) | <0.001 | — | — | — | — | 4.51 | 1.87 | 10.90 | 0.001 |
| IL28B rs8099917 genotype TT (vs. GT/GG) | 79 (96.3) | 38 (82.6) | 0.008 | 7.06 | 1.66 | 30.10 | 0.008 | 6.91 | 1.53 | 31.17 | 0.012 |
| APRI, mean ± SD | 1.8 ± 1.7 | 2.0 ± 1.7 | 0.706 | ||||||||
Data are presented as n (%) unless otherwise indicated.
ALT = alanine aminotransferase; APRI = aspartate aminotransferase (AST)/platelet ratio index; BMI = body mass index; CI = confidence interval; GGT = gamma-glutamyl transpeptidase; OR = odds ratio; RVR = rapid virological response; SVR = sustained virological response; SD = standard deviation.
a. Only covariates with p < 0.1 at univariate analysis were included in a multivariate model to determine independent determinants.
SVR rates stratified by IL28B SNP genotypes and RVR achievements
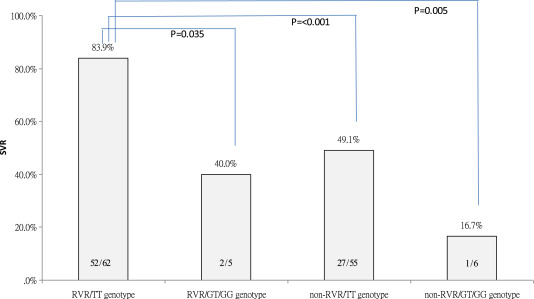
Because both IL28B genotype and RVR were determinant factors for SVR in the entire population, a group analysis of the influence of IL28B SNP on SVR by stratifying the achievement of RVR was performed. As plotted in Fig. 3 , the SVR rate was 83.9% in patients with RVR and favorable IL28B rs8099917 TT genotype, which was significantly higher than that in patients with the unfavorable IL28B (GT/GG)/with RVR (40.0%, p = 0.035), favorable IL28B (TT)/without RVR (49.1%, p < 0.001), or unfavorable IL28B (GT/GG)/without RVR (16.7%, p = 0.005). Other comparisons for each pair of groups did not reach statistical significance.
|
|
|
Figure 3. SVR rates stratified by IL28B rs8099917 genotype and RVR. The Arabic numerals at the base of the bar indicate the patient numbers. RVR = rapid virological response; SVR = sustained virological response. |
Predictors of SVR: subgroup analysis for either RVR or non-RVR patients
In a subsequent analysis, we analyzed the predictors associated with SVR limited to those who achieved an RVR: the multivariate analysis showed that only rs8099917 TT genotype predicted SVR (OR 7.80; 95% CI, 1.15–52.83; p = 0.035). Next, predictive factors associated with SVR were investigated for those who did not achieve an RVR: the multivariate analysis showed that only the achievement of cEVR was determined to be the predictor of SVR (OR 5.65; 95% CI, 1.12–28.52; p = 0.036).
Discussion
The results of this study extended what is already known about IL28B polymorphisms in patients with chronic hepatitis C by providing an insight into the relationship between IL28B rs8099917 genotype and virological response for HCV-1 infected patients treated with RGT. In our study, the combination of RVR and IL28B favorable genotype provided the best predictors of SVR, whereas the IL28B unfavorable genotype was a risk factor for relapse after treatment. For non-RVR patients, the achievement of cEVR, regardless of IL28B genotype, was the only prognosticator of SVR under the current RGT.
Low baseline viral loads predicted the attainment of an RVR in this study, which was not novel and was well investigated in volumes of studies in the past [19] . The impact of sex on treatment efficacy remains debatable. After adjusting for other factors including baseline viral loads, we found that male sex had a higher chance of RVR in the current study. The finding was in line with recent reports from Taiwan and Japan that male sex was one factor associated with good virological responses [15] ; [19] .
In general, favorable IL28B genotypes enhance early viral kinetics and SVR in HCV-1 patients [13] ; [15] ; [17] ; [18] ; [19] ; [21] ; unfavorable IL28B genotypes have been associated with slower viral decline and poor treatment efficacy, and this effect was particularly enhanced in patients who failed to achieve an RVR at Week 4 [17] ; [18] ; [19] ; [21] . We did not find an association between IL28B rs8099917 genotypes and RVR rate. The small sample size in the current study is one possible explanation. Despite this, we clearly demonstrated that patients who carried unfavorable IL28B genotypes had higher relapse rates under the current RGT (Fig. 2 ). Recently, Scherzer et al [22] also reported that relapse rates were lower among patients with a favorable IL28B genotype, and the extension of treatment duration was warranted for those who carried unfavorable alleles.
Regarding the achievement of an SVR, Model 1 (only baseline factors were included) showed that a lower baseline viral load and IL28B genotype affected the SVR rates. However, when RVR was taken into consideration in Model 2, the attainment of an RVR and IL28B genotype were predictors of SVR, but not baseline viral loads. Furthermore, for RVR patients (and all of them received an abbreviated 24-week treatment), IL28B genotype was the only predictor of SVR. When we analyzed the combination effect of these two prognosticators (IL28B and RVR) on SVR, as plotted in Fig. 3 , the SVR rate was 83.9% in patients with dual favorable factors, which yielded the highest treatment success rate compared to those patients who only had either one or no favorable factor. Previous studies have demonstrated that abbreviated therapy will not significantly compromise the likelihood of SVR among those who had achieved an RVR. Herein, we learned that this was not the case in the era of IL28B genotypic polymorphisms. Our important finding emphasized that an abbreviated 24-week therapy was suboptimal and should be avoided in RVR patients who had unfavorable IL28B genotype. We therefore suggested that IL28B genotypes be tested for all patients who had achieved an RVR. In line with our results, Liu et al [21] concluded that we could adopt the favorable L28B genotype plus RVR predicting rule to truncate treatment duration when patients are concerned about adverse events and increased medical costs.
Although the attainment of an RVR is the landmark of treatment success, only half of HCV-1 infected patients were able to achieve it in this study. More attention should be paid to patients who failed to reach the early goal. For non-RVR patients, we showed that only the achievement of cEVR and none of the baseline factors including IL28B genotype was determined to be the predictor of SVR. Our results prompted us to raise the important findings provided by Liu et al [20] regarding treatment effects of slow responders. They found that, for HCV-1 infected patients without RVR, the effects of IL28B genotype decreased when the on-treatment viral response (at Week 8 and Week 12) were taken into consideration; and IL28B polymorphisms play only a minor role in identifying optimal treatment duration in HCV-1 infected slow responders [20] . Although we could not totally ignore the discriminatory power of the IL28B genotypes in non-RVR patients [19] , our results highlighted that the achievement of cEVR was the most important factor to ensure treatment success for patients who failed to clear HCV during the first 4 weeks. Providing and maintaining the optimal dose of treatment within 12 weeks of treatment was pivotal for the attainment of cEVR, and every effort has to be made to achieve the second goal, cEVR, if patients failed to achieve RVR.
Previously, we did find that treatment adherence was an influential prognosticator among the treated cohort [24] . However, in this study, treatment adherence was not a prognosticator after the analyses. The reason may be that the discontinuation rate was low (3.9%) and the achievement rate of 80/80/80 adherent was already high (77.3%) in this series. In this intention-to-treat analysis, we certainly did not exclude patients who did not fulfill the 80/80/80 adherence rule, because the aim of this study was not an exploratory analysis of the influence of IL28B genotype on treatment responses. We believed that the study design we built here could provide information on useful prognosticators in our daily practice in a real-world setting. However, the major findings were not altered even if we excluded the 29 noncompliant patients who failed to meet the 80/80/80 adherent rule (data not shown).
The limitation of the current study is its retrospective nature, and the case number in certain subgroups might be too small. In addition, the completed histological data were unavailable in the current study, and we used the APRI score instead to represent the severity of liver disease [18] ; [19] ; [23] . The current study did not find any association between APRI score and treatment responses. Lastly, the definition of an RVR achievement in this study (undetectable HCV RNA, <15 IU/mL) was more stringent than the common one used in previous reports (HCV RNA <50 IU/mL). However, as one important finding we emphasized here was that an abbreviated 24-week therapy was suboptimal and should be avoided in RVR patients who had unfavorable IL28B genotype, we believed that using the stringent definition of RVR achievement was more relevant to our conclusion.
In conclusion, the results of this analysis confirmed the importance of IL28B genotype and on-treatment virological response in HCV-1 patients undergoing RGT. The combination of RVR and IL28B favorable genotype provided the best predictor of abbreviated 24-week treatment duration, whereas the IL28B unfavorable genotype was a risk factor of relapse after treatment. For HCV-1 infected patients without RVR, the achievement of cEVR, regardless of the IL28B genotype, was the only prognosticator of SVR under the current RGT.
Conflicts of interest
All authors declare no conflicts of interest.
Acknowledgments
The study was supported by a grant from Tungs Taichung MetroHarbor Hospital, Taichung, Taiwan (TTMHH-101R0021 ).
References
- [1] M. Omata, T. Kanda, M.L. Yu, O. Yokosuka, S.G. Lim, W. Jafri, et al.; APASL consensus statements and management algorithms for hepatitis C virus infection; Hepatol Int, 6 (2012), pp. 409–435
- [2] M.P. Manns, J.G. McHutchison, S.C. Gordon, V.K. Rustgi, M. Shiffman, R. Reindollar, et al.; Peginterferon-alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomized trial; Lancet, 358 (2001), pp. 958–965
- [3] M.W. Fried, M.L. Shiffman, K.R. Reddy, C. Smith, G. Marinos, F.L. Concales Jr., et al.; Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection; N Engl J Med, 347 (2002), pp. 975–982
- [4] S.J. Hadziyannis, H. Sette Jr., T.R. Morgan, V. Balan, M. Diago, P. Marcellin, et al.; Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose; Ann Intern Med, 140 (2004), pp. 346–355
- [5] F. Poordad, K.R. Reddy, P. Martin; Rapid virologic response: a new milestone in the management of chronic hepatitis C; Clin Infect Dis, 46 (2008), pp. 78–84
- [6] S. Zeuzem, D.R. Nelson, P. Marcellin; Dynamic evolution of therapy for chronic hepatitis C: how will novel agents be incorporated into the standard of care?; Antivir Ther, 13 (2008), pp. 747–760
- [7] P. Marcellin, E.J. Heathcote, A. Craxì; Which patients with genotype 1 chronic hepatitis C can benefit from prolonged treatment with the ‘accordion’ regimen?; J Hepatol, 47 (2007), pp. 580–587
- [8] S. Zeuzem, T. Berg, B. Moeller, H. Hinrichsen, S. Mauss, H. Wedemeyer, et al.; Expert opinion on the treatment of patients with chronic hepatitis C; J Viral Hepat, 16 (2009), pp. 75–90
- [9] A. Mangia, N. Minerva, D. Bacca, R. Cozzolongo, G.L. Ricci, V. Carretta, et al.; Individualized treatment duration for hepatitis C genotype 1 patients: a randomized controlled trial; Hepatology, 47 (2008), pp. 43–50
- [10] C.H. Liu, C.J. Liu, C.L. Lin, C.C. Liang, S.J. Hsu, S.S. Yang, et al.; Pegylated interferon-alpha-2a plus ribavirin for treatment-naive Asian patients with hepatitis C virus genotype 1 infection: a multicenter, randomized controlled trial; Clin Infect Dis, 47 (2008), pp. 1260–1269
- [11] M.L. Yu, C.Y. Dai, J.F. Huang, C.F. Chiu, Y.H. Yang, N.J. Hou, et al.; Rapid virological response and treatment duration for chronic hepatitis C genotype 1 patients: a randomized trial; Hepatology, 47 (2008), pp. 1884–1893
- [12] D.M. Jensen, T.R. Morgan, P. Marcellin, P.J. Pockros, K.R. Reddy, S.J. Hadziyannis, et al.; Early identification of HCV genotype 1 patients responding to 24 weeks peginterferon alpha-2a (40 kd)/ribavirin therapy; Hepatology, 43 (2006), pp. 954–960
- [13] D. Ge, J. Fellay, A.J. Thompson, J.S. Simon, K.V. Shianna, T.J. Urban, et al.; Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance; Nature, 461 (2009), pp. 399–401
- [14] D.L. Thomas, C.L. Thio, M.P. Martin, Y. Qi, D. Ge, C. O'Huigin, et al.; Genetic variation in IL28B and spontaneous clearance of hepatitis C virus; Nature, 461 (2009), pp. 798–801
- [15] Y. Tanaka, N. Nishida, M. Sugiyama, M. Kurosaki, K. Matsuura, N. Sakamoto, et al.; Genome-wide association of IL28B with response to pegylated interferon-[alpha] and ribavirin therapy for chronic hepatitis C; Nat Genet, 41 (2009), pp. 1105–1109
- [16] V. Suppiah, M. Moldovan, G. Ahlenstiel, T. Berg, M. Weltman, M.L. Abate, et al.; IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy; Nat Genet, 41 (2009), pp. 1100–1104
- [17] A.J. Thompson, A.J. Muir, M.S. Sulkowski, D. Ge, J. Fellay, K.V. Shianna, et al.; Interleukin- 28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus; Gastroenterology, 139 (2010), pp. 120–129
- [18] C.F. Huang, J.F. Huang, J.F. Yang, M.Y. Hsieh, Z.Y. Lin, S.C. Chen, et al.; Interleukin-28B genetic variants in identification of hepatitis C virus genotype 1 patients responding to 24 weeks peginterferon/ribavirin; J Hepatol, 56 (2012), pp. 34–40
- [19] C.F. Huang, M.L. Yeh, J.F. Huang, J.F. Yang, M.Y. Hsieh, Z.Y. Lin, et al.; Host interleukin-28B genetic variants versus viral kinetics in determining responses to standard-of-care for Asians with hepatitis C genotype 1; Antiviral Res, 93 (2012), pp. 239–244
- [20] C.H. Liu, C.C. Liang, C.J. Liu, T.C. Tseng, C.L. Lin, S.S. Yang, et al.; Interleukin 28B genetic polymorphisms play a minor role in identifying optimal treatment duration in HCV genotype 1 slow responders to pegylated interferon plus ribavirin; Antivir Ther, 17 (2012), pp. 1059–1067
- [21] C.H. Liu, C.C. Liang, C.J. Liu, T.C. Tseng, C.L. Lin, S.S. Yang, et al.; Interleukin 28B genetic polymorphisms and viral factors help identify HCV genotype-1 patients who benefit from 24-week pegylated interferon plus ribavirin therapy; Antivir Ther, 17 (2012), pp. 477–484
- [22] T.M. Scherzer, A.F. Stättermayer, M. Strasser, H. Laferl, A. Maieron, R. Stauber, et al.; Impact of IL28B on treatment outcome in hepatitis C virus G1/4 patients receiving response-guided therapy with peginterferon alpha-2a (40KD)/ribavirin; Hepatology, 54 (2011), pp. 1518–1526
- [23] C.T. Wai, J.K. Greenson, R.J. Fontana, J.D. Kalbfleisch, J.A. Marrero, H.S. Conjeevaram, et al.; A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C; Hepatology, 38 (2003), pp. 518–526
- [24] T.M. Chen, P.T. Huang, C.H. Lin, M.H. Tsai, L.F. Lin, C.C. Liu, et al.; Feasibility of individualized treatment for hepatitis C patients in the real world; J Gastroenterol Hepatol, 25 (2010), pp. 61–69
Document information
Published on 15/05/17
Submitted on 15/05/17
Licence: Other
Share this document
Keywords
claim authorship
Are you one of the authors of this document?