Recently an article published in Molecular Cell reveals the mechanism of a nuclear N6 -methyladenosine (m6 A) reader, the YTH domain-containing protein 1 (YTHDC1), in regulating pre-mRNA splicing [1] . Meanwhile, two additional articles published in Nature and Nature Chemical Biology report the first transcriptome-wide maps of N1 -methyladenosine (m1 A) at high resolution, suggesting a functional role for m1 A in translation regulation [2] and [3] .
m6 A reader YTHDC1 in pre-mRNA alternative splicing
m6 A is the most abundant endogenous mRNA modification, which is conserved across archea, bacteria, and eukaryotes [4] . Nonetheless, the importance of m6 A in mammals had been underappreciated for about 40 years until the discovery of its reversibility by an m6 A demethylase—fat mass and obesity-associated protein (FTO) [5] in 2011. Ever since, the widespread regulatory roles of m6 A have been unraveled through the transcriptome-wide mapping of m6 A modification [6] and [7] , the characterization of the second m6 A demethylase AlkB homolog 5 (ALKBH5) [8] and three subunits of m6 A methyltransferase complex (methyltransferase like 3, METTL3; METTL14; and Wilms tumor 1 associated protein, WTAP) [9] and [10] , and the functional studies of m6 A readers YTH domain family protein 1 (YTHDF1) and YTHDF2 in humans, which regulates m6 A methylated RNA stability [11] and translational efficiency [12] , respectively. In addition, m6 A in primary microRNAs can be recognized by another m6 A reader, the heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1), which consequently recruits DiGeorge syndrome critical region 8 (DGCR8) and DROSHA complex and promotes the maturation of microRNAs [13] and [14] .
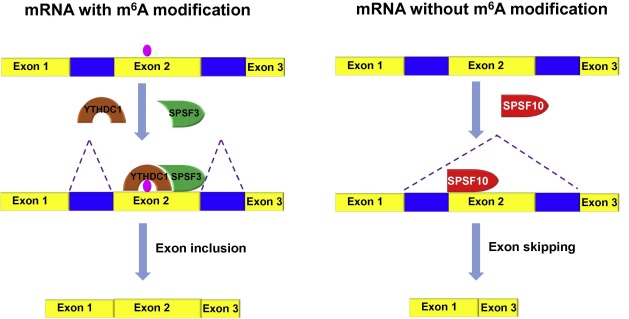
YTHDC1, as reflected by its name, contains the YTH domain that selectively binds to m6 A [15] . Unlike the other two cytoplasmic m6 A binding proteins YTHDF1 and YTHDF2, YTHDC1 is localized in YT bodies near the nuclear speckles [16] , supporting its association with pre-mRNA splicing. Xiao and colleagues [1] identified several YTHDC1 partners including five trans -acting splicing factors (serine/arginine-rich splicing factors; SRSF1/3/9/7/10) by tandem-affinity purification following by mass spectrometric analysis, suggesting the potential regulatory role of YTHDC1 in pre-mRNA splicing. To test such possibility, they measured the alternative splicing (AS) events using RNA-seq data upon knockdown of YTHDC1 and its potential SRSF partners in HeLa cells, respectively. Their findings indicate that YTHDC1 and SRSF3 facilitate exon inclusion, while SRSF10 promotes exon skipping; however, silencing of other SRSF proteins (SRSF1, SRSF7, and SRSF9) has no significant effect on AS events. Photoactivatable ribonucleoside crosslinking and immunoprecipitation (PAR-CLIP) sequencing shows that the targeted regions of YTHDC1, SRSF3, and SRSF10 are enriched in the coding sequences (CDS) and the 3′ untranslated regions (UTR). Through analyzing the targeted exons, they further confirmed the opposite roles of YTHDC1/SRSF3 and SRSF10 in AS regulation. The change of AS events on the transcripts targeted by both YTHDC1 and SRSF3 in HeLa cells with YTHDC1 or SRSF3 silenced shows similar features with that in METTL3-silenced HeLa cells, suggesting that YTHDC1 and SRSF3 co-regulates AS events in an m6 A-dependent manner.
Next, the authors set out to validate the interaction of YTHDC1 with either SRSF3 or SRSF10. PAR-CLIP data show that the YTHDC1 target regions are located closer to the binding sites of SRSF3 than those of SRSF10. In vivo and in vitro co-immunoprecipitation assay verifies that YTHDC1 directly interacts with SRSF3 and SRSF10 through the N-terminal of YTHDC1 and C-terminal of SRSF3 or SRSF10. The different AS events affected by YTHDC1/SRSF3 and SRSF10 prompts them to speculate that SRSF3 and SRSF10 might competitively bind to YTHDC1. Indeed they confirm the hypothesis using competing pull-down assays. The authors then examine whether YTHDC1 regulates localization of SRSF3 and SRSF10. Immunostaining assays show that silencing of YTHDC1 reduces SRSF3 but increases SRSF10 in nuclear speckle. Interestingly, this phenomenon can be rescued by complementation of wild-type YTHDC1, but not YTHDC1 mutant without m6 A binding ability, indicating that YTHDC1 regulates the subcellular localization of SRSF3 and SRSF10 in an m6 A-dependent manner. Further RNA binding assay shows that YTHDC1 deficiency disrupts the RNA binding of SRSF3 but enhances that of SRSF10, which can be complemented by wild-type YTHDC1, but not an m6 A-binding-defective variant. These results indicate that the impact of YTHDC1 on AS events relies on the presence of m6 A and the binding ability of YTHDC1 to methylated RNA.
Clearly, the comprehensive analysis presented by Xiao et al. reveals that m6 A reader YTHDC1 facilitates exon inclusion by recruiting RNA splicing factor SRSF3 but blocking SRSF10 for its access to the binding regions of its target mRNAs (Figure 1 ). Indeed, apart from YTHDC1, m6 A reader HNRNPA2B1 [14] and indirect m6 A reader HNRNPC [17] are both involved in RNA splicing. What roles do these proteins play in AS? Are there any other splicing factors regulated by m6 A? Does YTHDC1 play other regulatory role apart from splicing? These questions warrant further investigations.
|
|
|
Figure 1. A proposed model of pre-mRNA splicing regulated by YTHDC1Under the conditions that m6 A in pre-mRNA is recognized by YTHDC1, YTHDC1 recruits SRSF3 to promote exon inclusion; under the conditions that pre-mRNA does not contain m6 A or pre-mRNA with m6 A is not bound by YTHDC1, SRSF10 facilitates exon skipping. YTHDC1, YTH domain-containing protein 1; SRSF, serine/arginine-rich splicing factor. |
The reversible and dynamic m1 A methylome in eukaryotic mRNA
m1 A, another RNA adenosine methylation modification, has been identified in total RNA [18] , rRNA [19] , and tRNA [20] for decades. m1 A modification contains a methyl group on N1 (hydrogen bond receptor) to form the positive charge and disturbs Watson–Crick base pairs. Unlike m6 A, m1 A can cause both reverse transcription stops and read-throughs accompanied by mismatches. m1 A has been shown to affect the structure and function of tRNA and rRNA [21] and [22] . However, the presence and functions of m1 A in mRNA remain unknown.
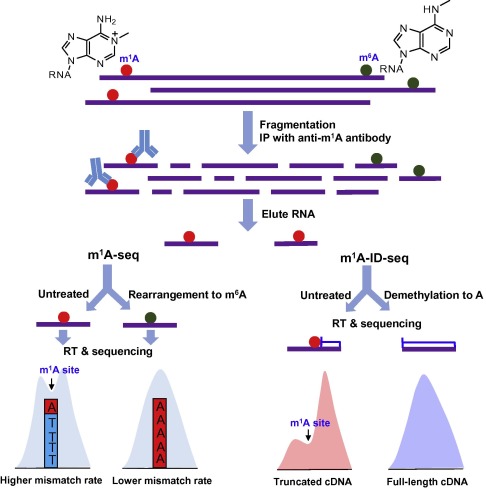
In the two recently-released papers, Dominissini et al. [2] and Li et al. [3] reported two transcriptome-wide sequencing methods (termed m1 A-seq and m1 A-ID-seq, respectively) to map m1 A in mRNA at high resolution (Figure 2 ). Their work reveals that m1 A is the second reversible and dynamic modification in eukaryotic mRNA. They firstly enrich m1 A-containing mRNA fragments from human or mouse cell lines by m1 A-specific antibody immunoprecipitation, and then take advantage of m1 A property in reverse transcription to improve the sequencing resolution, albeit later on the two groups employ different approaches for locating m1 A sites (Figure 2 ). As m1 A modification can be converted to m6 A in alkaline conditions (Dimroth rearrangement), Dominissini et al. treated a portion of precipitated m1 A-containing mRNA fragments with alkaline buffer to chemically rearrange m1 A to m6 A prior to cDNA synthesis. By comparing mismatch rates between treated and untreated samples, they located m1 A position within m1 A peaks, in which mutation rates are high in the treated sample but low in the untreated sample. In this way, they can achieve m1 A sequencing peaks at the resolution of 5–15 nucleotides (conserved m1 A sites in rRNA can be mapped at the resolution of one nucleotide) [2] (Figure 2 ). Different from Dominissini et al., Li et al. used Escherichia coli AlkB protein to demethylate m1 A to regular adenosine and performed cDNA synthesis with AMV reverse transcriptase to maximally confer cDNA truncations near m1 A sites. In this way, they achieved the m1 A map at the resolution of 55 nucleotides by comparing the m1 A peak features between the untreated and treated samples [3] (Figure 2 ). In fact, both strategies, based on mutations or truncations, sacrifice the sequencing signal and lose some sequence information near the modified sites, which make it difficult to obtain single-base resolution m1 A maps of high quality.
|
|
|
Figure 2. Schematic outline of m1 A-seq and m1 A-ID-seq In m1 A-seq, mismatch rates caused by m1 A (untreated sample) and m6 A (chemical rearrangement) were compared. In m1 A-ID-seq, cDNA truncations conferred by m1 A (untreated sample) were compared to full-length cDNA (demethylation to A). IP, immunoprecipitation; RT, reverse transcription. |
The relative abundance of m1 A in mammalian mRNA is much lower (m1 A/A: 0.015%–0.054% in cell lines and up to 0.16% in tissues) than that of m6 A (m6 A/A: 0.4%–0.6%) [2] . m1 A-seq identified 7154 m1 A peaks covering 4151 coding and 63 non-coding genes in humans [2] , whereas m1 A-ID-seq detected 901 m1 A peaks with high confidence in 600 human genes [3] . Both studies show that most of the identified transcripts contain only one m1 A peak. Unlike m6 A peaks that are enriched in the last transcribed exon [6] , [7] , [23] and [24] , m1 A peaks are highly enriched within 5’ UTR and near start codons.
According to the estimation of Dominissini and colleagues [2] , ∼20% genes contain a single m1 A. Through the deep analysis, they find that m1 A is associated with canonical and alternative translation initiation sites, as well as the first splice site. Therefore they presume that the first spicing reaction might guide m1 A deposition. m1 A prefers more structured regions with high GC content and low minimum free energy. It is of note that m1 A level and distribution pattern in mouse embryonic fibroblasts (MEFs) and mouse embryonic stem cells (mESCs) are comparable to those in human cell lines, suggesting an evolutionarily-conserved pattern of m1 A methylome. They also survey the influence of different stress conditions on m1 A, and find that the total level and peak number of m1 A can be reduced by glucose starvation but enhanced by heat shock, indicating the dynamic feature of m1 A under different physiological conditions. Given the close association of m1 A with the translation initiation sites, Dominissini and colleagues examine whether m1 A affects mRNA translation by using published ribosome profiling and proteomics data. Notably, m1 A-containing genes have higher translation efficiency and protein levels compared to non-m1 A-containing genes, implying that m1 A modification is correlated with elevated translation.
Meanwhile, Li and colleagues [3] studied the m1 A dynamics induced by H2 O2 treatment and serum starvation. They propose that m1 A may reside in a prominent motif with a GA-rich consensus. Similar with the aforementioned Nature paper, they state that m1 A prefers structured sequences with high GC content. It is notable that ALKBH3 (human ortholog of E. coli AlkB) is found to be able to demethylate m1 A in human mRNA, indicating that m1 A is a reversible modification and may play an important regulatory role on mRNA.
Collectively, the two studies by Dominissini and his colleagues [2] and Li and his colleagues [3] provide the first map of transcriptome-wide m1 A methylome and suggest new roles for m1 A: this reversible modification is enriched around start codon, dynamically regulated by stress conditions, and correlated with elevated translation. Although the two m1 A-seq techniques discussed here provide m1 A maps with relatively-high resolution compared to m6 A-seq method (at the resolution of ∼200 nucleotides), a big challenge is to develop single-base resolution methods for m6 A and for m1 A as well. Another challenge is to uncover the broader biological functions of m6 A and m1 A modifications. Future studies will focus on the identification and characterization of writer and reader proteins and functional roles of these two modifications. Given that m6 A as an RNA structure switch affects RNA–protein interaction [17] , the RNA structure changed by m1 A modification might also play certain functions. We expect more investigations to draw a more comprehensive picture of RNA modification story.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program; Grant No. 2014CB964900 ) and the National Natural Science Foundation of China (Grant Nos. 21432002 , 21372022 , and 21210003 ).
References
- [1] W. Xiao, S. Adhikari, U. Dahal, Y.S. Chen, Y.J. Hao, B.F. Sun, et al.; Nuclear m6 A reader YTHDC1 regulates mRNA splicing ; Mol Cell, 61 (2016), pp. 507–519
- [2] D. Dominissini, S. Nachtergaele, S. Moshitch-Moshkovitz, E. Peer, N. Kol, M.S. Ben-Haim, et al.; The dynamic N1 -methyladenosine methylome in eukaryotic messenger RNA ; Nature, 530 (2016), pp. 441–446
- [3] X. Li, X. Xiong, K. Wang, L. Wang, X. Shu, S. Ma, et al.; Transcriptome-wide mapping reveals reversible and dynamic N1 -methyladenosine methylome ; Nat Chem Biol, 12 (2016), pp. 311–316
- [4] Y. Yue, J. Liu, C. He; RNA N6 -methyladenosine methylation in post-transcriptional gene expression regulation ; Genes Dev, 29 (2015), pp. 1343–1355
- [5] G. Jia, Y. Fu, X. Zhao, Q. Dai, G. Zheng, Y. Yang, et al.; N6 -Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO ; Nat Chem Biol, 7 (2011), pp. 885–887
- [6] D. Dominissini, S. Moshitch-Moshkovitz, S. Schwartz, M. Salmon-Divon, L. Ungar, S. Osenberg, et al.; Topology of the human and mouse m6 A RNA methylomes revealed by m6 A-seq ; Nature, 485 (2012), pp. 201–206
- [7] K.D. Meyer, Y. Saletore, P. Zumbo, O. Elemento, C.E. Mason, S.R. Jaffrey; Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons; Cell, 149 (2012), pp. 1635–1646
- [8] G. Zheng, J.A. Dahl, Y. Niu, P. Fedorcsak, C.-M. Huang, C.J. Li, et al.; ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility; Mol Cell, 49 (2013), pp. 18–29
- [9] J. Liu, Y. Yue, D. Han, X. Wang, Y. Fu, L. Zhang, et al.; A METTL3–METTL14 complex mediates mammalian nuclear RNA N6 -adenosine methylation ; Nat Chem Biol, 10 (2014), pp. 93–95
- [10] X.L. Ping, B.F. Sun, L. Wang, W. Xiao, X. Yang, W.J. Wang, et al.; Mammalian WTAP is a regulatory subunit of the RNA N6 -methyladenosine methyltransferase ; Cell Res, 24 (2014), pp. 177–189
- [11] X. Wang, Z. Lu, A. Gomez, G.C. Hon, Y. Yue, D. Han, et al.; N6 -methyladenosine-dependent regulation of messenger RNA stability ; Nature, 505 (2014), pp. 117–120
- [12] X. Wang, B.S. Zhao, I.A. Roundtree, Z. Lu, D. Han, H. Ma, et al.; N6 -Methyladenosine modulates messenger RNA translation efficiency ; Cell, 161 (2015), pp. 1388–1399
- [13] C.R. Alarcón, H. Lee, H. Goodarzi, N. Halberg, S.F. Tavazoie; N6 -Methyladenosine marks primary microRNAs for processing ; Nature, 519 (2015), pp. 482–485
- [14] C.R. Alarcón, H. Goodarzi, H. Lee, X. Liu, S. Tavazoie, S.F. Tavazoie; HNRNPA2B1 is a mediator of m6 A-dependent nuclear RNA processing events ; Cell, 162 (2015), pp. 1299–1308
- [15] C. Xu, X. Wang, K. Liu, I.A. Roundtree, W. Tempel, Y. Li, et al.; Structural basis for selective binding of m6 A RNA by the YTHDC1 YTH domain ; Nat Chem Biol, 10 (2014), pp. 927–929
- [16] O. Nayler, A.M. Hartmann, S. Stamm; The ER repeat protein YT521-B localizes to a novel subnuclear compartment; J Cell Biol, 150 (2000), pp. 949–962
- [17] N. Liu, Q. Dai, G. Zheng, C. He, M. Parisien, T. Pan; N6 -Methyladenosine-dependent RNA structural switches regulate RNA–protein interactions ; Nature, 518 (2015), pp. 560–564
- [18] D. Dunn; The occurence of 1-methyladenine in ribonucleic acid; Biochim Biophys Acta, 46 (1961), pp. 198–200
- [19] R. Srivastava, K.P. Gopinathan; Ribosomal-RNA methylation in Mycobacterium smegmatis SN2 ; Biochem Int, 15 (1987), pp. 1179–1188
- [20] B. El Yacoubi, M. Bailly, V. de Crécy-Lagard; Biosynthesis and function of posttranscriptional modifications of transfer RNAs; Annu Rev Genet, 46 (2012), pp. 69–95
- [21] M. Helm, H. Brulé, F. Degoul, C. Cepanec, J.-P. Leroux, R. Giegé, et al.; The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA; Nucleic Acids Res, 26 (1998), pp. 1636–1643
- [22] C. Peifer, S. Sharma, P. Watzinger, S. Lamberth, P. Kötter, K.D. Entian; Yeast Rrp8p, a novel methyltransferase responsible for m1 A 645 base modification of 25S rRNA ; Nucleic Acids Res, 41 (2013), pp. 1151–1163
- [23] P.J. Batista, B. Molinie, J. Wang, K. Qu, J. Zhang, L. Li, et al.; m6 A RNA modification controls cell fate transition in mammalian embryonic stem cells ; Cell Stem Cell, 15 (2014), pp. 707–719
- [24] S. Ke, E.A. Alemu, C. Mertens, E.C. Gantman, J.J. Fak, A. Mele, et al.; A majority of m6 A residues are in the last exons, allowing the potential for 3′UTR regulation ; Genes Dev, 29 (2015), pp. 2037–2053
Document information
Published on 20/10/16
Licence: Other
Share this document
claim authorship
Are you one of the authors of this document?