Abstract
Cancer is a complex disease marked by uncontrolled cell growth and invasion. These processes are driven by the accumulation of genetic and epigenetic alterations that promote cancer initiation and progression. Contributing to genome changes are the regulation of oxidative stress and reactive species-induced damage to molecules and organelles. Redox regulation, metabolic plasticity, autophagy, and mitophagy play important and interactive roles in cancer hallmarks including sustained proliferation, activated invasion, and replicative immortality. However, the impact of these processes can differ depending on the signaling pathways altered in cancer, tumor type, tumor stage, and/or the differentiation state. Here, we highlight some of the representative studies on the impact of oxidative and nitrosative activities, mitochondrial bioenergetics, metabolism, and autophagy and mitophagy in the context of tumorigenesis. We discuss the implications of these processes for cellular activities in cancer for anti-cancer-based therapeutics.
Keywords
Autophagy; Cancer; Mitochondria; Mitophagy; Oxidative stress; Reactive species
As the name suggests, reactive oxygen species (ROS) are molecules that contain oxygen and are highly reactive. ROS include hydroxyl radical, hydrogen peroxide, and superoxide. The reaction of superoxide and the free radical nitric oxide also produces peroxynitrite, a potent oxidant. The molecules are produced by specific enzymatic pathways including the mitochondrial electron transport chain NOX/nicotinamide adenine dinucleotide phosphate oxidases and nitric oxide synthases [1], [2], [3], [4], [5], [6] and [7]. These reactive species can act as cell signaling molecules and also cause nonspecific posttranslational modification of proteins if domain-dependent control of their action is lost [8], [9], [10], [11], [12] and [13]. Under such circumstances, the irreversible modification of lipid, DNA, and proteins can accumulate in the cell and inactivate the biological function of these macromolecules as well as the organelles with which they are associated [14] and [15]. The maintenance of a redox homeostasis is then critical to both reductive and oxidative stress occurring when regulation of these pathways is lost [16]. Also, cancer initiation and progression are significantly impacted by redox signaling as well as redox stress [17].
Cellular metabolism is essential to generate adenosine triphosphate to provide the energy needed for multiple cellular functions. Such functions include DNA replication, transcription, translation, protein transport, assembly of multi-molecule complexes and organelles, cell mobility, and enzymatic reactions. In addition, the metabolites generated are building blocks for synthesis of DNA, RNA, and other essential cellular constituents. Metabolic programs are controlled at the levels of uptake of nutrients from extracellular space, glycolysis, and mitochondrial respiration. These metabolic functions can be regulated by reactive species and can also, in turn, regulate cellular redox status.
Autophagy and mitophagy are lysosome-mediated degradation of intracellular lipids, proteins, and organelles [18], [19], [20] and [21]. This degradation can serve to clear reactive species-induced damage to these molecules and cellular compartments. The processes are highly regulated by more than 30 proteins and many signaling pathways [21], which can include redox signaling itself as well as cellular metabolic programs [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32] and [33].
The integration of these cellular activities plays important roles in cancer initiation and progression and will be discussed in this review.
Reactive species, signaling, and stress in cancer initiation and progression
DNA damage by reactive species can be carcinogenic as represented by the link between cigarette smoking and increased risk of cancer [34], [35], [36] and [37]. Cigarette smoke contains reactive species such as nitric oxide, hydrogen peroxide, and peroxynitrite [34] and [35], and cigarette smokers exhibit increased oxidative damage as evidenced by elevated 8-hydroxy-2′deoxyguanosine (8-OHdG) levels [36] and [37]. The involvement of ROS in transformation mediated by oncogenes or tumor suppressor loss has also been demonstrated. For example, exogenous expression of oncogenic, constitutively active H-RasG12V or Myc leads to ROS-dependent transformation or mitogenic activities [38], [39], [40] and [41]. Downregulation of tumor suppressor genes such as p53 also leads to increased intracellular ROS, DNA oxidation, and mutation rate. Reactive nitrogen species also contribute to tumor growth as demonstrated by studies suggesting the importance of nitric oxide for cancer growth and tumor initiating cell maintenance [42], [43], [44], [45] and [46]. In converse, antioxidant-related drugs and molecules have been shown to inhibit tumor initiation. For example, the antioxidant N-acetylcysteine attenuates lymphomas in p53 knockout mice [47] and [48]. A transcription factor, critical for upregulation of antioxidant enzymes, nuclear factor (erythroid-derived 2) factor 2 (Nrf2), also attenuates cancer initiation similar to the antioxidant proteins it upregulates [49], [50], [51], [52], [53], [54], [55] and [56]. Together, these data suggest a pro-tumorigenic role for reactive species and a benefit for antioxidant-based therapies.
While reactive species can be pro-tumorigenic, their contribution to tumor biology is diverse and needs to be carefully considered prior to the development of novel therapeutic approaches. Evidence indicates that the role of ROS and antioxidants can differ depending on cell type or disease state. For example, mouse embryonic fibroblasts expressing endogenous K-RasG12V have lower levels of ROS as detected in the dichlorofluorescein fluorescence assay [57]. In this model, another indicator of oxidative stress, the glutathione (GSH) disulfide (oxidized GSH) to GSH (reduced glutathione; GSH) ratio, is also decreased in Nrf2-dependent fashion [57]. In tumor cells, high levels of antioxidant production through mechanisms such as upregulation of Nrf2 can provide survival advantages and resistance to chemotherapy [17], [56], [58], [59], [60], [61], [62] and [63]. Indeed, dual inhibition of the antioxidants glutathione and thioredoxin synergistically decreases tumor cell growth in vivo [167]. Overexpression of the anti-apoptotic factor Bcl2 also causes lymphomas in mice and humans without altering the rate of peroxide generation while attenuating oxidative damage to lipid membranes [64]. These studies highlight the complexity of the role of reactive species in cancer, which may vary depending on the genetic, epigenetic, and microenvironmental variation present in tumors. Thus, it may be difficult to make broad conclusions regarding the use of antioxidants for cancer therapy in the context of the diverse initiation and progression mechanisms of the disease [17], [55], [56] and [62].
Metabolic programming in cancer initiation and progression
Obesity increases the risk for various cancers, consistent with a close link of whole body metabolism to cancer predisposition [65] and [66]. At the cellular level, it has been long noted that metabolic programs in cancer cells differ from normal cells [67]. Recent studies identified diverse mechanisms of metabolic plasticity in cancer cells. These include increased glucose uptake in most tumors [68], [69], [70] and [71], elevated glycolytic intermediates due to the expression of the pyruvate kinase M2 isoform [72], [73], [74], [75] and [76], increased pentose phosphate pathway activities associated with transketolase isoform TKTL1 elevation [77] and [78], increased glutamine catabolism [79] and [80], and increased use of lactate as a fuel in selective tumors [81]. Signaling pathways and molecules, such as Akt and Myc that are known to play important roles in cancer, regulate the expression of glucose and glutamine transporters, glucose metabolism enzymes, glutamine metabolism, and mitochondrial biogenesis [82], [83], [84], [85], [86], [87], [88], [89], [90], [91] and [92]. Thus, it is becoming increasingly apparent that the pro-survival and pro-proliferative roles of oncogenic signals are strongly linked to changes in cellular metabolism and mitochondrial function. As an example, loss of the tumor suppressor p53 attenuates mitochondrial respiration and stimulates glycolysis with mechanisms including regulation of subunit I of cytochrome c oxidase, synthesis of cytochrome c oxidase 2, hexokinase 2, glucose transporters, phosphoglycerate mutase 1, and TP53-induced glycolysis and apoptosis regulator [93], [94], [95], [96], [97] and [98]. In human glioblastomas [99], [100] and [101] and acute myeloid leukemia [102], somatic mutations of isocitrate dehydrogenases alter metabolism by converting α-ketoglutarate to 2-hydroxyglutarate, which in turn leads to a hypermethylation phenotype [103] and [168]. The causal relationship of altered metabolism in tumorigenesis has also been suggested by the finding that germline mutations of succinate dehydrogenase and fumarate hydratase cause hereditary tumors, likely mediated by multifaceted mechanisms including altered gene expression, altered cell signaling, increased mutagenesis, or upregulation of hypoxia inducible factor 1 alpha (HIF1α) [104], [105], [106] and [107]. The data have led to increasing recognition that genetic mutations and metabolic changes in cancer are linked and required for the development and progression of the disease.
Autophagy and mitophagy in cancer initiation and progression
Autophagy senses cellular metabolic status, as well as various stress signals [29] and [108]. More than 30 proteins coordinate the autophagic processes, generating autophagosomes from essentially all membrane sources from the cell. Key signaling pathways include AMP-activated protein kinase (AMPK)-mammalian target of rapamycin (mTOR) pathways, Beclin-VPS34 complexes, ATG3, 4, 5, 6, 7, 8, 12, and 16 that are involved in autophagosomal formation, and adaptor proteins sequestosome 1 (SQSTM1)/p62 and NDP52 that recognize ubiquitinated targets. Perturbations of these pathways have been shown to contribute to tumorigenesis [109]. For example, mTOR inhibitors tuberous sclerosis complex-1/2 are tumor suppressors [110]. Beclin/ATG6 deficiency has been found to be associated with human breast, ovarian, and prostate tumors [111]; Beclin +/− mice develop tumors in endocrine tissues [112] and [113]. ATG4C, ATG5, and ATG7 genetic disruptions have also been found to develop liver adenomas or exhibit increased tumorigenesis in response to carcinogens in mice [114] and [115].
One regulator of autophagic flux associated with cancer is GABA(A) receptor-associated protein-like 1 (GABARAPL1). Lower levels of GABARAPL1 have been associated with poor outcome for liver and breast cancer patients [116] and [117], and GABARAPL1 is suggested to be a tumor suppressor through the inhibition of Wnt [118] and [119]. GABARAPL1 interacts with the Wnt/β-catenin signaling activator segment polarity protein disheveled homolog Dvl-2 and plays an important role in Dvl-2 degradation. Data indicate that knockdown of GABARAPL1 in breast cancer cells promotes proliferation and invasion in association with decreased autophagic flux and decreased lysosome numbers [120]. Although the requirement for Wnt in this effect is not known, Wnt signaling is well-established as a regulator of breast cancer initiation and metastasis [121] and [122], suggesting the attenuated GABARAPL1-mediated repression of Wnt signaling promotes breast cancer development and progression. Together, these data strongly suggest that a better understanding of the critical regulators of autophagy cellular transformation and tumor growth is important.
The above evidence indicates that autophagic deficits promote tumorigenesis as autophagy is required to guard against oxidative damage to the genome [23]. However, as multiple mechanisms are involved in both autophagy regulation and tumorigenesis, the connection between autophagy and tumor biology can be complex. For example, ablation of FIP200, a downstream target of mTOR inhibition and cofactor of ATG1-ATG13 activation, has been shown to inhibit mammary tumorigenesis [123]. In established tumors, autophagy may provide a survival advantage to the tumor cells in nutrient-deprived conditions and support chemoresistance [124]. Thus, autophagy inhibitors, such as chloroquine, have been tested in cancer therapy [125] and [126].
Mitophagy or autophagy of mitochondria is required to eliminate dysfunctional mitochondria to maintain appropriate metabolic and cell survival signals [32]. One key mediator of mitophagy linked to cancer is the putative tumor suppressor gene Parkin. Parkin is located at a chromosomal fragile site and loss is associated with tumors of the lung, breast, brain, ovary, pancreas, and colon [127], [128], [129], [130], [131], [132], [133] and [134]. In animal models, deletion of exon 3 of Parkin also promotes the development of spontaneous hepatic tumors [131]. The mechanisms through which Parkin acts as a tumor suppressor continue to be elucidated, but it is known that Parkin translocates to the mitochondria as a consequence of loss of membrane potential, leading to ubiquitination of mitochondrial proteins and recruitment of p62-LC3 and autophagosomes to the mitochondria [29], [32], [135], [136] and [137]. It regulates the ubiquitination of multiple mitochondrial proteins [138] with important targets being identified as the mitochondria fusion regulator Mitofusin 2 [139] and the mitochondria migration regulator mitochondrial rhoGTPase (Miro) [140]. Recent evidence suggesting the potential involvement of these Parkin targets in cancer further demonstrates the importance of mitophagy and mitochondrial function in cancer [141], [142], [143] and [144].
Parkin recruitment to promote mitophagy can be regulated by Bcl2/adenovirus E1B 19 kDa-interacting protein 3 (BNip3), and very recent evidence demonstrates that BNip3 integrates mitophagy and apoptosis signaling in cancer [145] and [146]. BNip3 utilizes its BH3 domain to inhibit pro-survival Bcl2 family members, and thereby activating apoptosis while also increasing mitophagy by binding autophagosomes through an LC3 interacting region. BNip3 loss has been associated with progression of breast cancer to metastasis through a mechanism thought to involve retention of dysfunctional mitochondria [147]. BNip3 loss prevents normal mitophagy, leading to elevated levels of ROS. In this breast cancer model system, increased ROS resulted in elevated levels of HIF1α and its target genes. The activation of HIF signaling resulted in increased glycolysis, consistent with a Warburg effect. Thus, the data directly linked loss of BNip3 regulated mitophagy to changes in metabolism promoting cancer progression. As BNip3 itself is a well-known HIF target gene, these data also demonstrate the complex interactions between the pathways regulating and being regulated by mitophagy [148].
On the other hand, in established tumors, an increased mitophagy associated with an increased autophagy may also provide a survival advantage to the tumor cells in nutrient deprived or hypoxic conditions and support chemoresistance. However, it is unclear whether increased mitophagy without increasing general autophagy occurs in advanced tumors, and whether specific inhibition of mitophagy sensitizes established tumors to chemotherapies.
Integration of reactive species, metabolic programs, and autophagy in cancer
The relationship between reactive species and metabolic programs and its role in cancer have been extensively studied. Reactive species modification of DNA, lipids, or protein clearly impacts cell metabolism and proliferation [149], [150] and [151]. Examples include the oxidative modification of mitochondrial DNA and mitochondrial proteins, as well as the induction of cell signaling and transcription pathways that are tumorigenic [41], [152] and [153]. Conversely, mitochondrial dysfunction plays a direct role in modulating cellular redox status [154] and [155] as well as generation of NADH and reduced GSH by the pentose phosphate pathway [153].
The relationship between reactive species and autophagy is also emerging as being important in cancer biology [156]. Reactive species regulation of autophagy has been demonstrated by thiol modification of ATG4 cysteines and signaling through the ataxia-telangiectasia mutated-liver kinase B1-AMPK-mTOR pathway [157] and [158]. Reactive species modification of Kelch-like ECH-associated protein 1 (KEAP1) leads to upregulation of the antioxidant regulating transcription factor Nrf2 and increased levels of autophagy adaptor protein SQSTM1/p62 [159]. Conversely, autophagy regulates KEAP1 levels and homeostasis of the KEAP1-Nrf2 pathway, thereby regulating cellular redox status [160].
The interactions among reactive stress, metabolism, and autophagy in cancer initiation and progression are complex. Strong evidence indicated these interactions in cancer. For example, the major nutrient-sensing Akt-AMPK-mTOR pathway is regulated by reactive stress and is a direct regulator of autophagy [158]. In pancreatic ductal adenocarcinoma cells, increased nuclear import of microphthalmia/transcription factor E family of transcription factors enhances autophagy-lysosomal catabolic function, maintains intracellular amino acid pools, and thereby supporting cell proliferation [161]. The tumor suppressor gene p53 has been shown to be activated by nutrient and oxidative stress while its activation also plays a role in metabolic homeostasis and autophagy by a variety of transcriptional as well as cytosolic mechanisms [162]. Recent studies also suggested that the covalent attachment of N-acetylglucosamine (O-GlcNAc) is not only involved in nutrient sensing and cancer metabolism [163], but may also play a role in regulating autophagy [164], [165] and [166]. These data demonstrate that there are multiple cross regulation mechanisms for ROS, autophagy, and metabolic signals in cancer.
Conclusion
Existing evidence supports the general concept that cancer initiation can be facilitated by changes in reactive species, autophagy, and metabolism. These changes may include increased reactive species modification of mitogenic signaling, reactive damage to DNA and proteins, decreased autophagic and mitophagic clearance of damaged macromolecules and organelles, and altered metabolic substrate availabilities and usages. Tumor cells with established genome mutations or rearrangements are highly dependent on metabolic plasticity to sustain proliferation, antioxidants, autophagy, and mitophagy to gain survival advantages. Cross-regulation of reactive species production and elimination, mitochondrial bioenergetics and glucose metabolism, and autophagy and mitophagy add additional complexity of the biology of tumorigenesis. Detailed or individualized mechanisms in specific tumors and specific stages of tumorigenesis and progression can be diverse and are still being intensively investigated. Studies on the regulation of mitochondrial bioenergetics, metabolism, autophagy and mitophagy, and how reactive species integrate these regulations in different cancers will continue to provide important insights into cancer biology and therapeutics.
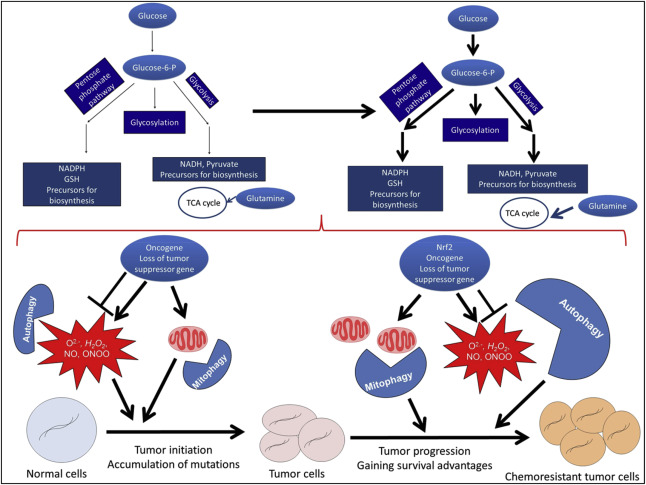
Normal cells are dependent on reactive species for cell signaling, mitochondria and glycolysis for energy, metabolites for biosynthesis, and autophagy for clearance of excessive or damaged macromolecules and organelles. Dysregulated reactive species generation and deficient autophagy contribute to tumorigenesis by enhancing mutagenesis and genome instability. Once tumors are established, tumor cells with upregulated autophagy and Nrf2-mediated antioxidant production may gain a survival advantage in low nutrient and hypoxic conditions. In addition, metabolic activities, such as mitochondrial biogenesis, glycolytic activities, and glutamine utilization, are upregulated to help sustain the energy demand for tumor growth. Understanding the coordination of these activities during tumorigenesis and progression is important for the prevention and management of cancer [[[#fig1|Fig. 1]]].
|
|
|
Fig. 1. Redox regulation, metabolic programming, autophagy, and mitophagy in cancer initiation and progression. |
Financial support and sponsorship
A.B. Hjelmeland is supported by CA1515122, the UAB Brain Tumor SPORE Career Development Award, and startup funds from the University of Alabama at Birmingham Department of Cell, Developmental and Integrative Biology. J. Zhang is supported by NIHR01-NS064090.
Conflicts of interest
There are no conflicts of interest.
References
- [1] S. Moncada, R.M. Palmer, E.A. Higgs; Nitric oxide: physiology, pathophysiology, and pharmacology; Pharmacol Rev, 43 (1991), pp. 109–142
- [2] R.G. Knowles, S. Moncada; Nitric oxide synthases in mammals; Biochem J, 298 (1994), pp. 249–258
- [3] P. Pacher, J.S. Beckman, L. Liaudet; Nitric oxide and peroxynitrite in health and disease; Physiol Rev, 87 (2007), pp. 315–424
- [4] B.G. Hill, B.P. Dranka, S.M. Bailey, J.R. Lancaster Jr., V.M. Darley-Usmar; What part of NO don't you understand? Some answers to the cardinal questions in nitric oxide biology; J Biol Chem, 285 (2010), pp. 19699–19704
- [5] M.P. Murphy, A. Holmgren, N.G. Larsson, B. Halliwell, C.J. Chang, B. Kalyanaraman, et al.; Unraveling the biological roles of reactive oxygen species; Cell Metab, 13 (2011), pp. 361–366
- [6] B. Kalyanaraman; Teaching the basics of redox biology to medical and graduate students: oxidants, antioxidants and disease mechanisms; Redox Biol, 8 (1) (2013), pp. 244–257
- [7] J.D. Lambeth, A.S. Neish; Nox enzymes and new thinking on reactive oxygen: a double-edged sword revisited; Annu Rev Pathol, 9 (2014), pp. 119–145
- [8] V.J. Thannickal, B.L. Fanburg; Reactive oxygen species in cell signaling; Am J Physiol Lung Cell Mol Physiol, 279 (2000), pp. L1005–L1028
- [9] H. Sauer, M. Wartenberg, J. Hescheler; Reactive oxygen species as intracellular messengers during cell growth and differentiation; Cell Physiol Biochem, 11 (2001), pp. 173–186
- [10] A. Landar, V.M. Darley-Usmar; Nitric oxide and cell signaling: modulation of redox tone and protein modification; Amino Acids, 2 (2003), pp. 313–321
- [11] S. Shiva, D. Moellering, A. Ramachandran, A.L. Levonen, A. Landar, A. Venkatraman, et al.; Redox signalling: from nitric oxide to oxidized lipids; Biochem Soc Symp, 71 (2004), pp. 107–120
- [12] M.L. Circu, T.Y. Aw; Reactive oxygen species, cellular redox systems, and apoptosis; Free Radic Biol Med, 48 (2010), pp. 749–762
- [13] A. Higdon, A.R. Diers, J.Y. Oh, A. Landar, V.M. Darley-Usmar; Cell signalling by reactive lipid species: new concepts and molecular mechanisms; Biochem J, 442 (2012), pp. 453–464
- [14] K. Hensley, K.A. Robinson, S.P. Gabbita, S. Salsman, R.A. Floyd; Reactive oxygen species, cell signaling, and cell injury; Free Radic Biol Med, 28 (2000), pp. 1456–1462
- [15] T. Finkel, N.J. Holbrook; Oxidants, oxidative stress and the biology of ageing; Nature, 408 (2000), pp. 239–247
- [16] E.S. Christians, I.J. Benjamin; Proteostasis and REDOX state in the heart; Am J Physiol Heart Circ Physiol, 302 (2012), pp. H24–H37
- [17] A. Glasauer, N.S. Chandel; Targeting antioxidants for cancer therapy; Biochem Pharmacol, 92 (2014), pp. 90–101
- [18] Z. Yang, D.J. Klionsky; Eaten alive: a history of macroautophagy; Nat Cell Biol, 12 (2010), pp. 814–822
- [19] Y. Chen, D.J. Klionsky; The regulation of autophagy – unanswered questions; J Cell Sci, 124 (2011), pp. 161–170
- [20] K. Wang, D.J. Klionsky; Mitochondria removal by autophagy; Autophagy, 7 (2011), pp. 297–300
- [21] Y. Feng, D. He, Z. Yao, D.J. Klionsky; The machinery of macroautophagy; Cell Res, 24 (2014), pp. 24–41
- [22] D.C. Rubinsztein, P. Codogno, B. Levine; Autophagy modulation as a potential therapeutic target for diverse diseases; Nat Rev Drug Discov, 11 (2012), pp. 709–730
- [23] E. White; Deconvoluting the context-dependent role for autophagy in cancer; Nat Rev Cancer, 12 (2012), pp. 401–410
- [24] J. Lee, S. Giordano, J. Zhang; Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling; Biochem J, 441 (2012), pp. 523–540
- [25] B.G. Hill, G.A. Benavides, J.R. Lancaster Jr., S. Ballinger, L. Dell'Italia, Z. Jianhua, et al.; Integration of cellular bioenergetics with mitochondrial quality control and autophagy; Biol Chem, 393 (2012), pp. 1485–1512
- [26] A.M. Choi, S.W. Ryter, B. Levine; Autophagy in human health and disease; N Engl J Med, 368 (2013), pp. 1845–1846
- [27] L. Murrow, J. Debnath; Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease; Annu Rev Pathol, 8 (2013), pp. 105–137
- [28] S. Giordano, V. Darley-Usmar, J. Zhang; Autophagy as an essential cellular antioxidant pathway in neurodegenerative disease; Redox Biol, 2 (2013), pp. 82–90
- [29] J. Zhang; Autophagy and mitophagy in cellular damage control; Redox Biol, 1 (2013), pp. 19–23
- [30] M. Dodson, V. Darley-Usmar, J. Zhang; Cellular metabolic and autophagic pathways: traffic control by redox signaling; Free Radic Biol Med, 63 (2013), pp. 207–221
- [31] A.L. Levonen, B.G. Hill, E. Kansanen, J. Zhang, V.M. Darley-Usmar; Redox regulation of antioxidants, autophagy, and the response to stress: implications for electrophile therapeutics; Free Radic Biol Med, 71 (2014), pp. 196–207
- [32] M. Redmann, M. Dodson, M. Boyer-Guittaut, V. Darley-Usmar, J. Zhang; Mitophagy mechanisms and role in human diseases; Int J Biochem Cell Biol, 53 (2014), pp. 127–133
- [33] D.B. Allison, L.H. Antoine, S.W. Ballinger, M.M. Bamman, P. Biga, V.M. Darley-Usmar, et al.; Aging and energetics' 'Top 40' future research opportunities 2010–2013; F1000Res, 3 (2014), p. 219
- [34] W.A. Pryor, K. Stone; Oxidants in cigarette smoke. Radicals, hydrogen peroxide, peroxynitrate, and peroxynitrite; Ann N Y Acad Sci, 686 (1993), pp. 12–27
- [35] J.P. Eiserich, V. Vossen, C.A. O'Neill, B. Halliwell, C.E. Cross, A. van der Vliet; Molecular mechanisms of damage by excess nitrogen oxides: nitration of tyrosine by gas-phase cigarette smoke; FEBS Lett, 353 (1994), pp. 53–56
- [36] S. Loft, H.E. Poulsen; Cancer risk and oxidative DNA damage in man; J Mol Med Berl, 74 (1996), pp. 297–312
- [37] A.J. Sasco, M.B. Secretan, K. Straif; Tobacco smoking and cancer: a brief review of recent epidemiological evidence; Lung Cancer, 45 (2004), pp. S3–S9
- [38] K. Irani, Y. Xia, J.L. Zweier, S.J. Sollott, C.J. Der, E.R. Fearon, et al.; Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts; Science, 275 (1997), pp. 1649–1652
- [39] K. Irani, P.J. Goldschmidt-Clermont; Ras, superoxide and signal transduction; Biochem Pharmacol, 55 (1998), pp. 1339–1346
- [40] O. Vafa, M. Wade, S. Kern, M. Beeche, T.K. Pandita, G.M. Hampton, et al.; c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability; Mol Cell, 9 (2002), pp. 1031–1044
- [41] F. Weinberg, R. Hamanaka, W.W. Wheaton, S. Weinberg, J. Joseph, M. Lopez, et al.; Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity; Proc Natl Acad Sci USA, 107 (2010), pp. 8788–8793
- [42] C.E. Eyler, Q. Wu, K. Yan, J.M. MacSwords, D. Chandler-Militello, K.L. Misuraca, et al.; Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2; Cell, 146 (2011), pp. 53–66
- [43] N. Charles, T. Ozawa, M. Squatrito, A.M. Bleau, C.W. Brennan, D. Hambardzumyan, et al.; Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells; Cell Stem Cell, 6 (2010), pp. 141–152
- [44] J.L. Heinecke, L.A. Ridnour, R.Y. Cheng, C.H. Switzer, M.M. Lizardo, C. Khanna, et al.; Tumor microenvironment-based feed-forward regulation of NOS2 in breast cancer progression; Proc Natl Acad Sci USA, 111 (2014), pp. 6323–6328
- [45] S.A. Glynn, B.J. Boersma, T.H. Dorsey, M. Yi, H.G. Yfantis, L.A. Ridnour, et al.; Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients; J Clin Invest, 120 (2010), pp. 3843–3854
- [46] M.A. Puglisi, C. Cenciarelli, V. Tesori, M. Cappellari, M. Martini, A.M. Di Francesco, et al.; High nitric oxide production, secondary to inducible nitric oxide synthase expression, is essential for regulation of the tumour-initiating properties of colon cancer stem cells; J Pathol, 236 (2015), pp. 479–490
- [47] A.A. Sablina, A.V. Budanov, G.V. Ilyinskaya, L.S. Agapova, J.E. Kravchenko, P.M. Chumakov; The antioxidant function of the p53 tumor suppressor; Nat Med, 11 (2005), pp. 1306–1313
- [48] T. Li, N. Kon, L. Jiang, M. Tan, T. Ludwig, Y. Zhao, et al.; Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence; Cell, 149 (2012), pp. 1269–1283
- [49] M. Ramos-Gomez, M.K. Kwak, P.M. Dolan, K. Itoh, M. Yamamoto, P. Talalay, et al.; Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice; Proc Natl Acad Sci USA, 98 (2001), pp. 3410–3415
- [50] K. Iida, K. Itoh, Y. Kumagai, R. Oyasu, K. Hattori, K. Kawai, et al.; Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis; Cancer Res, 64 (2004), pp. 6424–6431
- [51] C. Xu, M.T. Huang, G. Shen, X. Yuan, W. Lin, T.O. Khor, et al.; Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2; Cancer Res, 66 (2006), pp. 8293–8296
- [52] M.S. Yates, M.K. Kwak, P.A. Egner, J.D. Groopman, S. Bodreddigari, T.R. Sutter, et al.; Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole; Cancer Res, 66 (2006), pp. 2488–2494
- [53] K.L. Cheung, J.H. Lee, T.O. Khor, T.Y. Wu, G.X. Li, J. Chan, et al.; Nrf2 knockout enhances intestinal tumorigenesis in Apc(min/+) mice due to attenuation of anti-oxidative stress pathway while potentiates inflammation; Mol Carcinog, 53 (2014), pp. 77–84
- [54] T.O. Khor, M.T. Huang, A. Prawan, Y. Liu, X. Hao, S. Yu, et al.; Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer; Cancer Prev Res (Phila), 1 (2008), pp. 187–191
- [55] A. Lau, N.F. Villeneuve, Z. Sun, P.K. Wong, D.D. Zhang; Dual roles of Nrf2 in cancer; Pharmacol Res, 58 (2008), pp. 262–270
- [56] M.B. Sporn, K.T. Liby; NRF2 and cancer: the good, the bad and the importance of context; Nat Rev Cancer, 12 (2012), pp. 564–571
- [57] G.M. DeNicola, F.A. Karreth, T.J. Humpton, A. Gopinathan, C. Wei, K. Frese, et al.; Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis; Nature, 475 (2011), pp. 106–109
- [58] T. Shibata, T. Ohta, K.I. Tong, A. Kokubu, R. Odogawa, K. Tsuta, et al.; Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy; Proc Natl Acad Sci USA, 105 (2008), pp. 13568–13573
- [59] P. Zhang, A. Singh, S. Yegnasubramanian, D. Esopi, P. Kombairaju, M. Bodas, et al.; Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth; Mol Cancer Ther, 9 (2010), pp. 336–346
- [60] A. Ooi, J.C. Wong, D. Petillo, D. Roossien, V. Perrier-Trudova, D. Whitten, et al.; An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma; Cancer Cell, 20 (2011), pp. 511–523
- [61] J. Adam, E. Hatipoglu, L. O'Flaherty, N. Ternette, N. Sahgal, H. Lockstone, et al.; Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling; Cancer Cell, 20 (2011), pp. 524–537
- [62] E. Kansanen, S.M. Kuosmanen, H. Leinonen, A.L. Levonen; The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer; Redox Biol, 1 (2013), pp. 45–49
- [63] H. Satoh, T. Moriguchi, J. Takai, M. Ebina, M. Yamamoto; Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis; Cancer Res, 73 (2013), pp. 4158–4168
- [64] D.M. Hockenbery, Z.N. Oltvai, X.M. Yin, C.L. Milliman, S.J. Korsmeyer; Bcl-2 functions in an antioxidant pathway to prevent apoptosis; Cell, 75 (1993), pp. 241–251
- [65] E.E. Calle, R. Kaaks; Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms; Nat Rev Cancer, 4 (2004), pp. 579–591
- [66] E.E. Calle, C. Rodriguez, K. Walker-Thurmond, M.J. Thun; Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults; N Engl J Med, 348 (2003), pp. 1625–1638
- [67] O. Warburg, F. Wind, E. Negelein; The metabolism of tumors in the body; J Gen Physiol, 8 (1927), pp. 519–530
- [68] S.S. Gambhir, J. Czernin, J. Schwimmer, D.H. Silverman, R.E. Coleman, M.E. Phelps; A tabulated summary of the FDG PET literature; J Nucl Med, 42 (2001), pp. 1S–93S
- [69] S.S. Gambhir; Molecular imaging of cancer with positron emission tomography; Nat Rev Cancer, 2 (2002), pp. 683–693
- [70] D.A. Mankoff, J.F. Eary, J.M. Link, M. Muzi, J.G. Rajendran, A.M. Spence, et al.; Tumor-specific positron emission tomography imaging in patients: [18F] fluorodeoxyglucose and beyond; Clin Cancer Res, 13 (2007), pp. 3460–3469
- [71] W.A. Flavahan, Q. Wu, M. Hitomi, N. Rahim, Y. Kim, A.E. Sloan, et al.; Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake; Nat Neurosci, 16 (2013), pp. 1373–1382
- [72] S. Mazurek, C.B. Boschek, F. Hugo, E. Eigenbrodt; Pyruvate kinase type M2 and its role in tumor growth and spreading; Semin Cancer Biol, 15 (2005), pp. 300–308
- [73] H.R. Christofk, M.G. Vander Heiden, M.H. Harris, A. Ramanathan, R.E. Gerszten, R. Wei, et al.; The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth; Nature, 452 (2008), pp. 230–233
- [74] S. Mazurek; Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells; Int J Biochem Cell Biol, 43 (2011), pp. 969–980
- [75] D. Anastasiou, Y. Yu, W.J. Israelsen, J.K. Jiang, M.B. Boxer, B.S. Hong, et al.; Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis; Nat Chem Biol, 8 (2012), pp. 839–847
- [76] M.A. Iqbal, V. Gupta, P. Gopinath, S. Mazurek, R.N. Bamezai; Pyruvate kinase M2 and cancer: an updated assessment; FEBS Lett, 588 (2014), pp. 2685–2692
- [77] S. Langbein, M. Zerilli, A. Zur Hausen, W. Staiger, K. Rensch-Boschert, N. Lukan, et al.; Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted; Br J Cancer, 94 (2006), pp. 578–585
- [78] M. Földi, E. Stickeler, L. Bau, O. Kretz, D. Watermann, G. Gitsch, et al.; Transketolase protein TKTL1 overexpression: a potential biomarker and therapeutic target in breast cancer; Oncol Rep, 17 (2007), pp. 841–845
- [79] R.J. DeBerardinis, A. Mancuso, E. Daikhin, I. Nissim, M. Yudkoff, S. Wehrli, et al.; Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis; Proc Natl Acad Sci USA, 104 (2007), pp. 19345–19350
- [80] C. Yang, J. Sudderth, T. Dang, R.M. Bachoo, J.G. McDonald, R.J. DeBerardinis; Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling; Cancer Res, 69 (2009), pp. 7986–7993
- [81] P. Sonveaux, T. Copetti, C.J. De Saedeleer, F. Végran, J. Verrax, K.M. Kennedy, et al.; Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis; PLoS One, 7 (2012), p. e33418
- [82] A.D. Kohn, S.A. Summers, M.J. Birnbaum, R.A. Roth; Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation; J Biol Chem, 271 (1996), pp. 31372–31378
- [83] M.R. Calera, C. Martinez, H. Liu, A.K. Jack, M.J. Birnbaum, P.F. Pilch; Insulin increases the association of Akt-2 with Glut4-containing vesicles; J Biol Chem, 273 (1998), pp. 7201–7204
- [84] D.R. Plas, S. Talapatra, A.L. Edinger, J.C. Rathmell, C.B. Thompson; Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology; J Biol Chem, 276 (2001), pp. 12041–12048
- [85] A.L. Edinger, C.B. Thompson; Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake; Mol Biol Cell, 13 (2002), pp. 2276–2288
- [86] J.C. Rathmell, C.J. Fox, D.R. Plas, P.S. Hammerman, R.M. Cinalli, C.B. Thompson; Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival; Mol Cell Biol, 23 (2003), pp. 7315–7328
- [87] J.T. Barata, A. Silva, J.G. Brandao, L.M. Nadler, A.A. Cardoso, V.A. Boussiotis; Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells; J Exp Med, 200 (2004), pp. 659–669
- [88] R.L. Elstrom, D.E. Bauer, M. Buzzai, R. Karnauskas, M.H. Harris, D.R. Plas, et al.; Akt stimulates aerobic glycolysis in cancer cells; Cancer Res, 64 (2004), pp. 3892–3899
- [89] C.V. Dang, J.W. Kim, P. Gao, J. Yustein; The interplay between MYC and HIF in cancer; Nat Rev Cancer, 8 (2008), pp. 51–56
- [90] D.R. Wise, R.J. DeBerardinis, A. Mancuso, N. Sayed, X.Y. Zhang, H.K. Pfeiffer, et al.; Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction; Proc Natl Acad Sci USA, 105 (2008), pp. 18782–18787
- [91] R.J. DeBerardinis, T. Cheng; Qs next: the diverse functions of glutamine in metabolism, cell biology and cancer; Oncogene, 29 (2010), pp. 313–324
- [92] E. Anso, A.R. Mullen, D.W. Felsher, J.M. Matés, R.J. Deberardinis, N.S. Chandel; Metabolic changes in cancer cells upon suppression of MYC; Cancer Metab, 1 (2013), p. 7
- [93] S.P. Mathupala, C. Heese, P.L. Pedersen; Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53; J Biol Chem, 272 (1997), pp. 22776–22780
- [94] S. Okamura, C.C. Ng, K. Koyama, Y. Takei, H. Arakawa, M. Monden, et al.; Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system; Oncol Res, 11 (1999), pp. 281–285
- [95] F. Schwartzenberg-Bar-Yoseph, M. Armoni, E. Karnieli; The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression; Cancer Res, 64 (2004), pp. 2627–2633
- [96] S. Matoba, J.G. Kang, W.D. Patino, A. Wragg, M. Boehm, O. Gavrilova, et al.; p53 regulates mitochondrial respiration; Science, 312 (2006), pp. 1650–1653
- [97] K. Bensaad, A. Tsuruta, M.A. Selak, M.N. Vidal, K. Nakano, R. Bartrons, et al.; TIGAR, a p53-inducible regulator of glycolysis and apoptosis; Cell, 126 (2006), pp. 107–120
- [98] T. Hitosugi, L. Zhou, S. Elf, J. Fan, H.B. Kang, J.H. Seo, et al.; Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth; Cancer Cell, 22 (2012), pp. 585–600
- [99] D.W. Parsons, S. Jones, X. Zhang, J.C. Lin, R.J. Leary, P. Angenendt, et al.; An integrated genomic analysis of human glioblastoma multiforme; Science, 321 (2008), pp. 1807–1812
- [100] H. Yan, D.D. Bigner, V. Velculescu, D.W. Parsons; Mutant metabolic enzymes are at the origin of gliomas; Cancer Res, 69 (2009), pp. 9157–9159
- [101] H. Yan, D.W. Parsons, G. Jin, R. McLendon, B.A. Rasheed, W. Yuan, et al.; IDH1 and IDH2 mutations in gliomas; N Engl J Med, 360 (2009), pp. 765–773
- [102] E.R. Mardis, L. Ding, D.J. Dooling, D.E. Larson, M.D. McLellan, K. Chen, et al.; Recurring mutations found by sequencing an acute myeloid leukemia genome; N Engl J Med, 361 (2009), pp. 1058–1066
- [103] L. Dang, D.W. White, S. Gross, B.D. Bennett, M.A. Bittinger, E.M. Driggers, et al.; Cancer-associated IDH1 mutations produce 2-hydroxyglutarate; Nature, 462 (2009), pp. 739–744
- [104] S. Zhao, Y. Lin, W. Xu, W. Jiang, Z. Zha, P. Wang, et al.; Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha; Science, 324 (2009), pp. 261–265
- [105] P.J. Pollard, J.J. Brière, N.A. Alam, J. Barwell, E. Barclay, N.C. Wortham, et al.; Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations; Hum Mol Genet, 14 (2005), pp. 2231–2239
- [106] M.A. Selak, S.M. Armour, E.D. MacKenzie, H. Boulahbel, D.G. Watson, K.D. Mansfield, et al.; Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase; Cancer Cell, 7 (2005), pp. 77–85
- [107] C. Bardella, P.J. Pollard, I. Tomlinson; SDH mutations in cancer; Biochim Biophys Acta, 1807 (2011), pp. 1432–1443
- [108] J. Zhang; Teaching the basics of autophagy and mitophagy to redox biologists – mechanisms and experimental approaches; Redox Biol, 4 (2015), pp. 242–259
- [109] S. Lorin, A. Hamaï, M. Mehrpour, P. Codogno; Autophagy regulation and its role in cancer; Semin Cancer Biol, 23 (2013), pp. 361–379
- [110] K. Inoki, M.N. Corradetti, K.L. Guan; Dysregulation of the TSC-mTOR pathway in human disease; Nat Genet, 37 (2005), pp. 19–24
- [111] X.H. Liang, S. Jackson, M. Seaman, K. Brown, B. Kempkes, H. Hibshoosh, et al.; Induction of autophagy and inhibition of tumorigenesis by beclin 1; Nature, 402 (1999), pp. 672–676
- [112] X. Qu, J. Yu, G. Bhagat, N. Furuya, H. Hibshoosh, A. Troxel, et al.; Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene; J Clin Invest, 112 (2003), pp. 1809–1820
- [113] Z. Yue, S. Jin, C. Yang, A.J. Levine, N. Heintz; Beclin 1, an autophagy gene essential for early embryonic development, is a haploin sufficient tumor suppressor; Proc Natl Acad Sci USA, 100 (2003), pp. 15077–15082
- [114] G. Mariño, N. Salvador-Montoliu, A. Fueyo, E. Knecht, N. Mizushima, C. López-Otín; Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3; J Biol Chem, 282 (2007), pp. 18573–18583
- [115] A. Takamura, M. Komatsu, T. Hara, A. Sakamoto, C. Kishi, S. Waguri, et al.; Autophagy-deficient mice develop multiple liver tumors; Genes Dev, 25 (2011), pp. 795–800
- [116] A. Berthier, S. Seguin, A.J. Sasco, J.Y. Bobin, G. De Laroche, J. Datchary, et al.; High expression of gabarapl1 is associated with a better outcome for patients with lymph node-positive breast cancer; Br J Cancer, 102 (2010), pp. 1024–1031
- [117] C. Liu, Y. Xia, W. Jiang, Y. Liu, L. Yu; Low expression of GABARAPL1 is associated with a poor outcome for patients with hepatocellular carcinoma; Oncol Rep, 31 (2014), pp. 2043–2048
- [118] C. Gao, W. Cao, L. Bao, W. Zuo, G. Xie, T. Cai, et al.; Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation; Nat Cell Biol, 12 (2010), pp. 781–790
- [119] Y. Zhang, F. Wang, L. Han, Y. Wu, S. Li, X. Yang, et al.; GABARAPL1 negatively regulates Wnt/ß-catenin signaling by mediating Dvl2 degradation through the autophagy pathway; Cell Physiol Biochem, 27 (2011), pp. 503–512
- [120] M. Boyer-Guittaut, L. Poillet, Q. Liang, E. Bôle-Richard, X. Ouyang, G.A. Benavides, et al.; The role of GABARAPL1/GEC1 in autophagic flux and mitochondrial quality control in MDA-MB-436 breast cancer cells; Autophagy, 10 (2014), pp. 986–1003
- [121] J.L. Green, J. La, K.W. Yum, P. Desai, L.W. Rodewald, X. Zhang, et al.; Paracrine Wnt signaling both promotes and inhibits human breast tumor growth; Proc Natl Acad Sci USA, 110 (2013), pp. 6991–6996
- [122] A.S. Cleary, T.L. Leonard, S.A. Gestl, E.J. Gunther; Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers; Nature, 508 (2014), pp. 113–117
- [123] H. Wei, S. Wei, B. Gan, X. Peng, W. Zou, J.L. Guan; Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis; Genes Dev, 25 (2011), pp. 1510–1527
- [124] J.Y. Guo, B. Xia, E. White; Autophagy-mediated tumor promotion; Cell, 155 (2013), pp. 1216–1219
- [125] F. Janku, D.J. McConkey, D.S. Hong, R. Kurzrock; Autophagy as a target for anticancer therapy; Nat Rev Clin Oncol, 8 (2011), pp. 528–539
- [126] N. Chen, V. Karantza; Autophagy as a therapeutic target in cancer; Cancer Biol Ther, 11 (2011), pp. 157–168
- [127] S.R. Denison, F. Wang, N.A. Becker, B. Schüle, N. Kock, L.A. Phillips, et al.; Alterations in the common fragile site gene Parkin in ovarian and other cancers; Oncogene, 22 (2003), pp. 8370–8378
- [128] R. Cesari, E.S. Martin, G.A. Calin, F. Pentimalli, R. Bichi, H. McAdams, et al.; Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27; Proc Natl Acad Sci USA, 100 (2003), pp. 5956–5961
- [129] M.C. Picchio, E.S. Martin, R. Cesari, G.A. Calin, S. Yendamuri, T. Kuroki, et al.; Alterations of the tumor suppressor gene Parkin in non-small cell lung cancer; Clin Cancer Res, 10 (2004), pp. 2720–2724
- [130] F. Wang, S. Denison, J.P. Lai, L.A. Philips, D. Montoya, N. Kock, et al.; Parkin gene alterations in hepatocellular carcinoma; Genes Chromosom Cancer, 40 (2004), pp. 85–96
- [131] M. Fujiwara, H. Marusawa, H.Q. Wang, A. Iwai, K. Ikeuchi, Y. Imai, et al.; Parkin as a tumor suppressor gene for hepatocellular carcinoma; Oncogene, 27 (2008), pp. 6002–6011
- [132] S. Veeriah, B.S. Taylor, S. Meng, F. Fang, E. Yilmaz, I. Vivanco, et al.; Somatic mutations of the Parkinsons disease-associated gene PARK2 in glioblastoma and other human malignancies; Nat Genet, 42 (2010), pp. 77–82
- [133] C.W. Yeo, F.S. Ng, C. Chai, J.M. Tan, G.R. Koh, Y.K. Chong, et al.; Parkin pathway activation mitigates glioma cell proliferation and predicts patient survival; Cancer Res, 72 (2012), pp. 2543–2553
- [134] X. Sun, M. Liu, J. Hao, D. Li, Y. Luo, X. Wang, et al.; Parkin deficiency contributes to pancreatic tumorigenesis by inducing spindle multipolarity and misorientation; Cell Cycle, 12 (2013), pp. 1133–1141
- [135] S.M. Jin, R.J. Youle; PINK1- and Parkin-mediated mitophagy at a glance; J Cell Sci, 125 (2012), pp. 795–799
- [136] I. Novak; Mitophagy: a complex mechanism of mitochondrial removal; Antioxid Redox Signal, 17 (2012), pp. 794–802
- [137] W.X. Ding, X.M. Yin; Mitophagy: mechanisms, pathophysiological roles, and analysis; Biol Chem, 393 (2012), pp. 547–564
- [138] S.A. Sarraf, M. Raman, V. Guarani-Pereira, M.E. Sowa, E.L. Huttlin, S.P. Gygi, et al.; Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization; Nature, 496 (2013), pp. 372–376
- [139] Y. Chen, G.W. Dorn; 2nd. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria; Science, 340 (2013), pp. 471–475
- [140] X. Wang, D. Winter, G. Ashrafi, J. Schlehe, Y.L. Wong, D. Selkoe, et al.; PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility; Cell, 147 (2011), pp. 893–906
- [141] Y. Lou, R. Li, J. Liu, Y. Zhang, X. Zhang, B. Jin, et al.; Mitofusin-2 over-expresses and leads to dysregulation of cell cycle and cell invasion in lung adenocarcinoma; Med Oncol, 32 (2015), p. 132
- [142] B. Jin, G. Fu, H. Pan, X. Cheng, L. Zhou, J. Lv, et al.; Anti-tumour efficacy of mitofusin-2 in urinary bladder carcinoma; Med Oncol, 28 (2011), pp. S373–S380
- [143] G.E. Zhang, H.L. Jin, X.K. Lin, C. Chen, X.S. Liu, Q. Zhang, et al.; Anti-tumor effects of Mfn2 in gastric cancer; Int J Mol Sci, 14 (2013), pp. 13005–13021
- [144] J. Rehman, H.J. Zhang, P.T. Toth, Y. Zhang, G. Marsboom, Z. Hong, et al.; Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer; FASEB J, 26 (2012), pp. 2175–2186
- [145] Y. Lee, H.Y. Lee, R.A. Hanna; Gustafsson ÅB. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes; Am J Physiol Heart Circ Physiol, 301 (2011), pp. H1924–H1931
- [146] S.C. Choe, A. Hamacher-Brady, N.R. Brady; Autophagy capacity and sub-mitochondrial heterogeneity shape Bnip3-induced mitophagy regulation of apoptosis; Cell Commun Signal, 13 (2015), p. 37
- [147] A.H. Chourasia, K. Tracy, C. Frankenberger, M.L. Boland, M.N. Sharifi, L.E. Drake, et al.; Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis; EMBO Rep, 16 (2015), pp. 1145–1163
- [148] R.K. Bruick; Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia; Proc Natl Acad Sci USA, 97 (2000), pp. 9082–9087
- [149] P.A. Kramer, V.M. Darley-Usmar; The emerging theme of redox bioenergetics in health and disease; Biomed J, 38 (2015), pp. 294–300
- [150] B. Halliwell; Cell culture, oxidative stress, and antioxidants: avoiding pitfalls; Biomed J, 37 (2014), pp. 99–105
- [151] Y. Xu, S.Y. Qian; Anti-cancer activities of omega -6 polyunsaturated fatty acids; Biomed J, 37 (2014), pp. 112–119
- [152] M. Valko, C.J. Rhodes, J. Moncol, M. Izakovic, M. Mazur; Free radicals, metals and antioxidants in oxidative stress-induced cancer; Chem Biol Interact, 160 (2006), pp. 1–40
- [153] R.A. Cairns, I.S. Harris, T.W. Mak; Regulation of cancer cell metabolism; Nat Rev Cancer, 11 (2011), pp. 85–95
- [154] M. Brandon, P. Baldi, D.C. Wallace; Mitochondrial mutations in cancer; Oncogene, 25 (2006), pp. 4647–4662
- [155] M.P. Murphy; How mitochondria produce reactive oxygen species; Biochem J, 417 (2009), pp. 1–13
- [156] L. Poillet-Perez, G. Despouy, R. Delage-Mourroux, M. Boyer-Guittaut; Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy; Redox Biol, 4 (2015), pp. 184–192
- [157] R. Scherz-Shouval, E. Shvets, E. Fass, H. Shorer, L. Gil, Z. Elazar; Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4; EMBO J, 26 (2007), pp. 1749–1760
- [158] A. Alexander, S.L. Cai, J. Kim, A. Nanez, M. Sahin, K.H. MacLean, et al.; ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS; Proc Natl Acad Sci USA, 107 (2010), pp. 4153–4158
- [159] A. Jain, T. Lamark, E. Sjøttem, K.B. Larsen, J.A. Awuh, A. Øvervatn, et al.; p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription; J Biol Chem, 285 (2010), pp. 22576–22591
- [160] M. Komatsu, H. Kurokawa, S. Waguri, K. Taguchi, A. Kobayashi, Y. Ichimura, et al.; The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1; Nat Cell Biol, 12 (2010), pp. 213–223
- [161] R.M. Perera, S. Stoykova, B.N. Nicolay, K.N. Ross, J. Fitamant, M. Boukhali, et al.; Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism; Nature, 524 (2015), pp. 361–365
- [162] F. Kruiswijk, C.F. Labuschagne, K.H. Vousden; p53 in survival, death and metabolic health: a lifeguard with a licence to kill; Nat Rev Mol Cell Biol, 16 (2015), pp. 393–405
- [163] Z. Ma, K. Vosseller; Cancer metabolism and elevated O-GlcNAc in oncogenic signaling; J Biol Chem, 289 (2014), pp. 34457–34465
- [164] S.A. Marsh, P.C. Powell, L.J. Dell'italia, J.C. Chatham; Cardiac O-GlcNAcylation blunts autophagic signaling in the diabetic heart; Life Sci, 92 (2013), pp. 648–656
- [165] B. Guo, Q. Liang, L. Li, Z. Hu, F. Wu, P. Zhang, et al.; O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation; Nat Cell Biol, 16 (2014), pp. 1215–1226
- [166] W.Y. Wani, M. Boyer-Guittaut, M. Dodson, J. Chatham, V. Darley-Usmar, J. Zhang; Regulation of autophagy by protein post-translational modification; Lab Invest, 95 (2015), pp. 14–25
- [167] I.S. Harris, A.E. Treloar, S. Inoue, M. Sasaki, C. Gorrini, K.C. Lee, et al.; Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression; Cancer Cell, 27 (2015), pp. 211–222
- [168] S. Turcan, D. Rohle, A. Goenka, L.A. Walsh, F. Fang, E. Yilmaz, et al.; IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype; Nature, 483 (2012), pp. 479–483
Document information
Published on 20/10/16
Licence: Other
Share this document
Keywords
claim authorship
Are you one of the authors of this document?