Highlights
- Concentration and half-lives of soluble NO donors are in accurate predictors of NO.
- Many environmental factors influence the ultimate level of NO cells experience.
- The actual NO cells experience should be monitored throughout in vitro experiments.
Abstract
Applying soluble nitric oxide (NO) donors is the most widely used method to expose cells of interest to exogenous NO. Because of the complex equilibria that exist between components in culture media, the donor compound and NO itself, it is very challenging to predict the dose and duration of NO cells actually experience. To determine the actual level of NO experienced by cells exposed to soluble NO donors, we developed the CellNO Trap, a device that allows continuous, real-time monitoring of the level of NO adherent cells produce and/or experience in culture without the need to alter cell culturing procedures. Herein, we directly measured the level of NO that cells grown in the CellNO Trap experienced when soluble NO donors were added to solutions in culture wells and we characterized environmental conditions that effected the level of NO in in vitro culture conditions. Specifically, the dose and duration of NO generated by the soluble donors S-nitroso-N-acetylpenicillamine (SNAP), S-nitrosoglutathione (GSNO), S-nitrosocysteine (CysNO) and the diazeniumdiolate diethyltriamine (DETA/NO) were investigated in both phosphate buffered saline (PBS) and cell culture media. Other factors that were studied that potentially affect the ultimate NO level achieved with these donors included pH, presence of transition metals (ion species), redox level, presence of free thiol and relative volume of media. Then murine smooth muscle cell (MOVAS) with different NO donors but with the same effective concentration of available NO were examined and it was demonstrated that the cell proliferation ratio observed does not correlate with the half-lives of NO donors characterized in PBS, but does correlate well with the real-time NO profiles measured under the actual culture conditions. This data demonstrates the dynamic characteristic of the NO and NO donor in different biological systems and clearly illustrates the importance of tracking individual NO profiles under the actual biological conditions.
Graphical abstract
Keywords
Direct NO measurement ; In vitro cell culture ; Complete media ; Soluble NO donors
1. Introduction
Nitric oxide (NO) is a very common immune regulator [1] , critical neurotransmitter [2] and potent vasodilator [3] . NO deficiency is closely related to chronic cardiovascular diseases such as hypertension, coronary heart diseases, and arterial thrombotic disorders [4] . To restore physiologically useful levels of NO, NO releasing drugs (NO donors) have been widely used for more than one hundred years. Currently GSNO has been clinically used to prevent thrombosis [5] . Additionally SNAP derivatives have also shown considerable potential to maintain vascular tone [6] and [7] . However, caution needs to be exercised when using NO as a drug, since NO potentially brings about fatal side-effect such as shock [8] .
Many experiments have shown that NO donors exhibited different potencies in different circumstances [9] , [10] , [11] and [12] . Although half-lives of NO donors are commonly used to predict the level of NO achieved and the time frame over which cells experience NO while using these soluble donors, the potencies of NO releasing chemicals are difficult to predict. The underlying issue with inaccurate predictability of the potency of NO donors lies in the poorly characterized NO levels actually achieved in the complex and changing environments of biological conditions. It is of great importance to directly understand how much actual NO cells experience rather than making assumptions regarding approximated NO levels based only on the concentration and chemical properties of the parent NO donors when trying to understand the role NO plays in cellular behavior.
Some critical limitations associated with using soluble NO donors that need to be recognized include: (1) the NO donors analytical concentration does not equal to NO level that cells or tissues experience; (2) NO level that cells actually experience is influenced by species present within the buffer solutions and culture media and by cells and tissues; (3) the NO donor treatment time may not represent the true NO exposure time; (4) NO donors biological effects should not be considered as equal to the effect of NO.
Considering the high diffusivity and reactivity of NO, Lancaster [13] proposed the following expression (Eq. (1)) to represent the concentration of NO at any given site and given time point under physiological conditions:
|
|
( 1) |
where v represents the rate of NO generation at a specific site and time point, is the diffusion of NO according to Ficks law, and is the consumption of NO due to auto-oxidation. Eq. (1) demonstrates that the ultimate NO level is determined by the rates of NO generation, consumption and the distance between the NO source and site of interest when the system is non-homogeneous (i.e., layer of cells in a culture dish). The concentration of the NO donor can affect the generation rate and greatly affects the ultimate NO level but is not equal to the analytical concentration of the NO donor. Biological systems are complicated, where different species within the biological system can react with NO and complicate the consumption rate of NO, greatly influencing the ultimate NO level. NO itself has a very short half-life. The half-life of NO varies from less than a second to minutes depending on its concentration and specific environment [14] and [15] . This means that the duration of NO is primarily determined by the generation process. Once the generation process is over, there will be no further NO exposure (assuming other intermediate NO sources such as S-nitrosothiols are not formed). NO donors consist of parent compounds that contain various functional groups which remain after the release of NO and further complicate the environment under study. Additionally some reactions that affects cellular response may not be directly through NO such as direct S-NO transfer [9] , which can easily be confused with the effect of NO.
To fully understand the roles of NO and the factors that may affect NO levels when using soluble NO donors, there is a great need to understand how much NO is actually present at the cell layer. Previously, our laboratory developed the CellNO Trap, a chemiluminescence-based device for directly monitoring NO levels within solutions and NO levels that cells produce during the entire duration of experiments in real-time [16] . Herein, we used this same device to directly measure and analyze the NO generation profiles of different commonly used NO donor compounds (SNAP, GSNO, CysNO, and DETA/NO) during in vitro culturing. Different factors that influence these profiles including cell culture conditions, pH/CO2 , free thiol levels, oxidative stress and solution volume were examined in detail with the CellNO Trap. A demonstration of using this device to illustrate the potencies of different NO donors in inhibiting MOVAS cell proliferation to different degrees was introduced. This data indicates that the NO generation profiles of all NO donors investigated are very dynamic. Real-time NO data is a powerful tool to help explain observed biological data regarding potencies of NO donors used in experiments.
2. Materials and methods
2.1. Chemicals and cells
Silicone elastomer base & curing agent (Sylgard® 184) were ordered from Dow Corning Co. (Midland, MI). Penicillin-streptomycin (pen/strep), G418 disulfate, collagen I, N-acetyl-D,L-penicillamine, acetic anhydride, calcein-AM, hydrogen peroxide, di-tert-butyl peroxide, and cyclam were obtained from Sigma-Aldrich (St. Louis, MO). Tert-butyl nitrite was purchased from Acros Organics, (Pittsburgh, PA). Gelatin was obtained from Bio-rad (Hercules, CA). Pyridine was purchased from EMD Chemical Inc. (Darmstadt, Germany). Ethidium bromide, Click-iT® EdU assay kit and Hoechst dye were purchased from Invitrogen (Grand Island, NY). Smooth muscle cell line MOVAS, Dulbeccos modified eagle medium (DMEM), fetal bovine serum (FBS), were all purchased from ATCC (Manassas, VA).
2.2. CellNO Trap device manufacturing
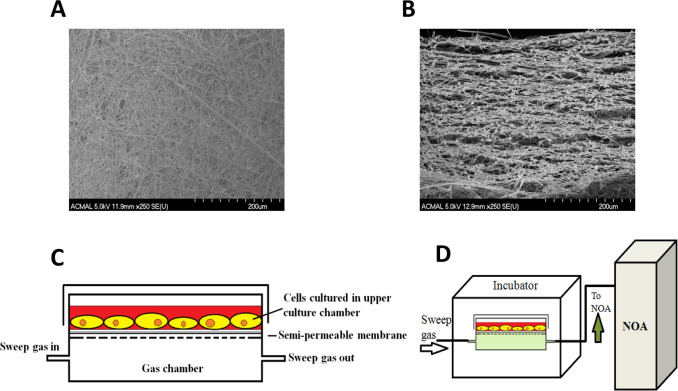
Detailed fabrication and characterization of the CellNO Trap can be found elsewhere [16] . Briefly, the semipermeable membrane for the CellNO Trap was manufactured through top treating glass fiber filter paper by manually casting 5% Sylgard® hexanes solution (v/v) for 3 times (72 μl/cm2 /cast) and air-drying. Then membrane was placed into 60 °C oven for overnight heat to stiffen the polymer. The surface and cross-section of the polymer treated foam-like micro structure of the membrane are shown in Fig. 1 A and B by SEM. The membrane was cut according to chamber frames and attached in between the two-chamber system by dropping toluene to temporally dissolve the plastic frame to adhere the membrane. The final device is shown in Fig. 1 C. For cell culture experiments, the device was sterilized by ethylene oxide and then top treated with 20 μg/ml collagen I solution for 2 h, or top coated with 1 mg/ml dopamine solution (10 mM Tris buffer, pH=8.5) for overnight, sterilized by ethylene oxide and treated by 2 mg/ml gelatin solution (dissolve in PBS) for 1 h. Air-dried device will be ready for cell culture.
|
|
|
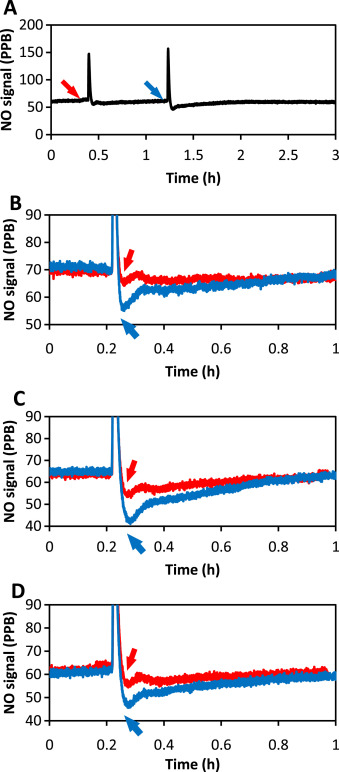
Fig. 1. Illustration of the CellNo Trap experimental set-up. (A) SEM image of the surface the semi-permeable membrane; (B) SEM of a cross-section of the semi-permeable membrane showing the foam-like structure which allows NO to pass through and provides a surface upon which cells can grow; (C) Illustration of the two-chamber NO measurement system; (D) real-time measurement of NO that cells experience using chemiluminescence detection. |
2.3. SNAP synthesis and quantification
S-nitroso-N-acetylpenicillamine crystals were synthesized by dissolving 200 mg of N-acetylpenicillamine (NAP) in 5 ml methanol along with sonication. Two to three milliliter of HCl and 100 μl of concentrated H2 SO4 were slowly added in order to acidify the solution. The acidified NAP solution was combined with 144.9 mg NaNO2 and vortex-mixed avoiding light till all the sodium nitrite was dissolved. After dark green/red color was gradually developed (SNAP solution), the solution was ice-cooled for 45 min. SNAP crystals were collected by rotary evaporation and vacuum filtration, and washed by diH2 O repeatedly for 3 times and air-dried. SNAP content was tested by injecting known mass percent of SNAP solution into excessive triiodide solution (both with and without acidified sulfanilamide) according to Yang et al.[17] .
2.4. GSNO synthesis and quantification
GSNO synthesis was adapted from Hart et al. [18] , and all the synthetic procedures were shielded from light. Briefly, 1.54 g of GSH was dissolved within 6.52 ml 1 M HCl and stirred on ice; 0.345 g of NaNO2 dissolved in 1 ml water was dropped gently into the GSH solution and to allow for reaction for 10 min; then 710 μl newly prepared 10 N NaOH was carefully added to neutralize the reaction system to pH between 3 and 4; the end product was aliquoted and stored in −80 °C freezer until use. The produced GSNO concentration should be close to 500 mM. Before each use, one vial was taken out to measure the concentration by UV–vis spectrometry at 335 nm, and the extinction coefficient used was 0.92 mM−1 cm−1 .
2.5. CysNO synthesis and quantification
CysNO synthesis was accomplished through nitrosating acidified cysteine (Cys). Cysteine was dissolved in 1 M HCl (final concentration 0.2 M) and stirred on ice on the magnetic plate, protected from light. Equimolar amounts of NaNO2 was carefully pipetted into the Cys solution and allowed to react for 10 min. Freshly prepared 10 N NaOH was carefully titrated into the CysNO solution until solution pH was between 3 and 4.The end product was aliquoted and stored in −80 °C until used. Before each use, one vial was taken out to measure the concentration with UV–vis spectrometry at 543 nm (ε=16.8 M−1 cm−1 ).
2.6. Cell culture and proliferation assays
Mouse smooth muscle cells (MOVAS) were cultured in DMEM media, with 10% FBS, 1% pen/strep, and 0.2 mg/ml G418 in 37 °C 5% CO2 incubator. Cells were seeded onto the cover-slip at an initial density of 10,000 cell/cm2 then placed into 6-well plate for culture, or cells were seeded directly into the surface of the CellNO Trap. After overnight culturing to allow cell recovery, the specific NO donor was added by freshly dissolving donor stock solutions into the culture media to 2X specific concentrations and gently adding equal volume of this 2X donor media solution to the volume of culturing media containing cells. Immediately before using all the NO donors, the concentration of the different NO donor stocks were determined by UV–vis spectrometry. Then culturing of cells continued for different durations, with a 24 h maximum time frame. Immediately after the designated treatment time ended, proliferating cells were labeled by using EdU imaging kit. In brief, DNA synthesis was labeled by EdU (5-ethynyl-2′-deoxyuridine) incorporation, and this incorporation is visualized by alkyne-azide (EdU provides alkyne; Alexa Fluor® dye provides azide) reaction, which was performed after 3.7% formaldehyde fixation and 0.5% Triton X-100 permeabilization. Nuclei were then stained by Hoechst. Proliferating cells were imaged under Olympus BX51 microscope and cell number was quantified by ImageJ.
2.7. Real-time NO measurement through chemiluminescence NOA
Before measurement, Sievers 280i Nitric Oxide Analyzer (GE Instruments, Boulder, CO) was zero calibrated by using sampling gas (either N2 gas, atmospheric air, or air within the incubator according to different needs) and calibration gas calibrated by using 45 PPM NO standard (Air Liquid Healthcare America Corp. Plumsteadville, PA) using 200 ml/min flow rate. Then the NOA calibration constant was determined by injecting 100 μl of 500 μM NaNO2 standard to stirring acidified Lugol solution. NO flux through the CellNO Trap was measured according to our previous published method [16] . In Brief, the CellNO Trap was connected to the NOA. Data recording was initiated before adding the donor solutions into the device to let the background signal stabilize. Then 37 °C pre-warmed bathing solution with specified concentrations of the NO donor of interest, with or without other chemicals such as free thiols or extra acid, were freshly prepared and quickly added to CellNO Trap for measurement (as shown in Fig. 1 D).
2.8. Statistical analysis
All NO measurement experiments were independently run 3 times unless specified otherwise. Cell proliferation and cell number experiments were independently repeated in triplicate with 3 samples for each treatment. Error bars represent sample standard deviations among each repeat. The data was analyzed by either student t -student test or one-way analysis of variance (ANOVA). All statistical assays were achieved through R programming unless specific notification.
3. Results and discussion
3.1. Definition and quantification of cellular NO level
Schmidt et al. [19] presented a straightforward method to calculate NO concentration from soluble NO donors by considering the NO generation rate of donors and the auto-oxidation of NO by oxygen. The primary limitation of this method is the calculation is only applicable to solutions with simple composition (where NO consumption routes are limited), making its application to real biological systems largely inaccurate. Herein, we introduce a method to measure NO flux (in pmol cm−2 s or 10−10 mol cm−2 min−1 ), which can be empirically measured by using the CellNO Trap without making assumptions regarding NO generation and consumption pathways or rates.
As illustrated in Fig. 1 , the upper chamber serves as a conventional cell culture environment to which drugs (including NO donors) can be directly applied to cells using established cell culture protocols. While NO is present in the cell environment, NO diffuses in all directions, and most importantly, through the semipermeable membrane into the lower chamber. NO in the lower chamber can be detected via chemiluminescence without influencing activities in the upper chamber. Once a solution with a specific NO donor is applied in the upper chamber, the actual NO level (dose and duration) at the surface of the cell layer of this particular donor can be tracked in real-time.
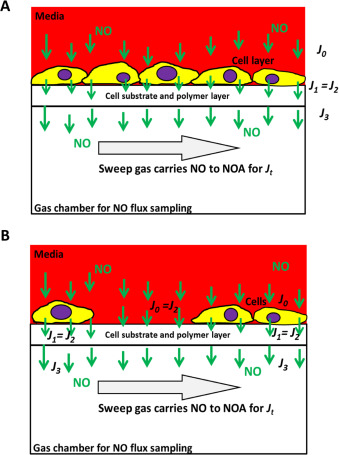
The key question in describing the NO levels cells experience is what this measured flux (or NO level) means to cells cultured in the device. To illustrate this, two models were shown below. First, assume a tightly attached intact mono layer of cell is cultured onto the device membrane to obtain the cell sheet model (see Fig. 2 A):
|
|
|
Fig. 2. NO quantification in the confluent mono cell layer model (A) and the nonconfluent cell culture model (B). |
- Suppose J0 is NO flux entering the cell;
J1 is the NO flux exiting the cell;
- J2 is the NO flux entering the semipermeable membrane;
- J3 is the NO flux exiting the semipermeable membrane;
- and Jt is the NO signal measured by the NOA.
NO will be consumed through each interface such as J0 to J1 . This decay is represented as Jc , so:
|
|
( 2) |
Predicting measured NO flux (Jt ) at time (t ) relative to J0 or J1 will allow the actual NO level around cells to be known. Because cells tightly attach to the membrane, J1 and J2 were considered to be equal, such that:
|
|
( 3) |
Also since the sampling process by a sweep gas in the lower chamber is a continuous and very fast process, it is considered that immediately after the NO travels through the membrane, the signal can be detected by the NOA, regardless of whether N2 or ambient air is used as the sweep gas, such that:
|
|
( 4) |
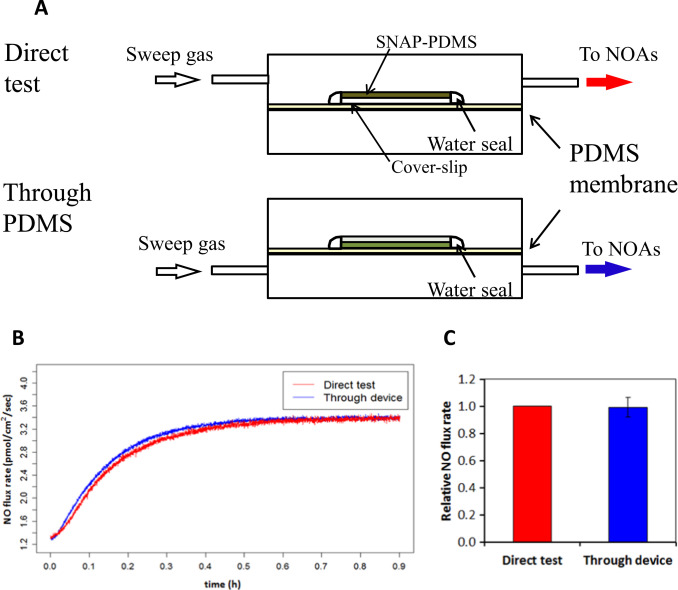
We designed an experiment to investigate how much NO consumption occurs between J2 and J3 . The highly controllable NO releasing polymer SNAP-PDMS was cast on a cover-slip and top coated with RTV-3140 PDMS layer. This SNAP-PDMS film releases NO at a certain constant rate at specific temperature (due to thermal degradation) [20] . By measuring the NO flux when the polymer layer faces up from the upper chamber, the original NO flux coming out of the polymer (Jp) was obtained and facing down and sampling from the bottom chamber, the NO diffusing across the membrane (JTr) was detected (Fig. 3 A). A membrane specific NO permeability η was defined as
|
|
( 5) |
|
|
|
Fig. 3. Evaluation of the cross-membrane signal dampening of NO. Panel (A) shows the experimental design of the evaluation of the NO cross-membrane signal dampening. Panel (B) illustrates real-time NO flux data of both Jp and JTr ; while panel (C) shows the comparison of direct measurement and cross-membrane measurement of NO. The average NO flux (n=5) at steady-state (defined as the NO flux from time 60–70 min after polymer was applied for measurement, where the NO signal became almost stable) was calculated; the NO flux at steady-state measured by direct measurement was normalized to itself and the corresponding cross-membrane NO flux was normalized to this value; P >0.9 according to paired t -test. |
Result (Fig. 3 B) shows that direct NO measurement and cross-membrane measurement produced almost identical NO releasing profiles. After 5 replicate measurements, the steady state NO flux values were averaged. Values were applied to paired t -test ( Fig. 3 C). Data showed that no detectable difference exists between the two sets of measurements using these two methods. This implies that the relative NO permeability η can be considered 100%, meaning there is negligible signal loss between Jp and JTr , i.e. from J2 to J3 , indicating:
|
|
( 6) |
In this model, from Eqs. (3) , (4) and (6) , it is reasonable to use Jt to represent J1 . So the NO level that was measured by the NOA is equivalent to the NO flux coming from the cell sheet.
In the second model, if cells are not confluent (assume k % is the fractional area covered in Fig. 2 B), since Eqs. (4) and (6) are still valid here, it is easy to obtain the following relation to represent Jt :
|
|
( 7) |
In addition, while the cell coverage ratio is very low and the reaction occurred in the cells is negligible (i.e. a very small Jc ), such that Jt and J0 might be considered as equal in the very low cell density model. This NO flux Jt is used to reflect the dose and duration of NO that cells actually experience when NO donors are introduced in solution to the culture media.
3.2. NO release levels
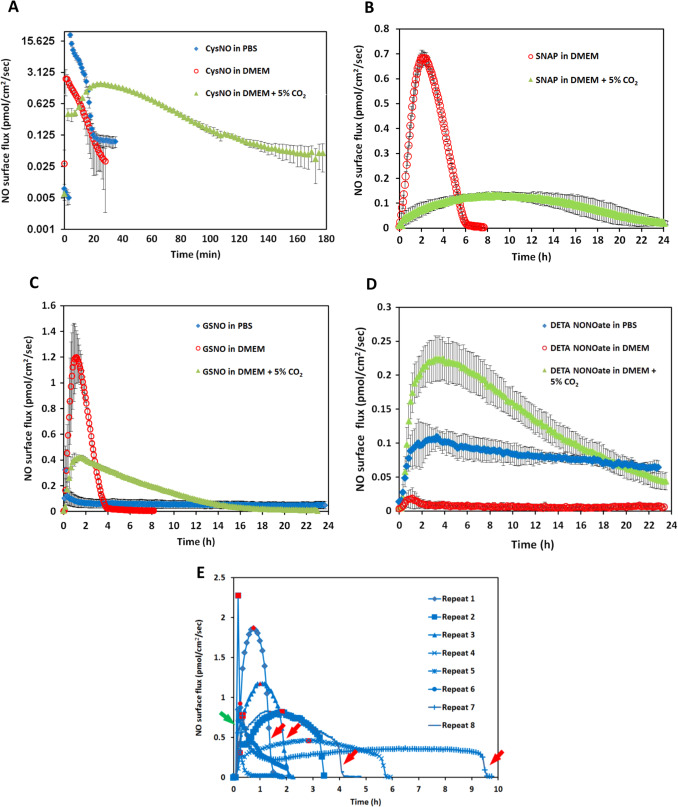
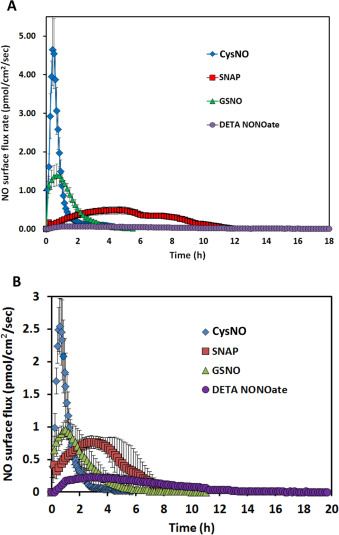
The NO release levels (the level of NO and the time duration over which this level is experienced) of four different commonly used NO donors were examined by using the CellNO Trap device. In order to elucidate the NO level in real biological conditions, all the experiments were run at 37 °C in PBS buffer or complete cell culture DMEM media. The same effective concentration of NO donors (i.e. such that the same moles of NO were in theory generated) was used for direct comparison. Biologically irrelevant concentrations were avoided, so 50 μM of donor was used in most of the following experiments. Considering the reported NO donors’ half-lives [13] and [21] , it is estimated that these soluble NO donor systems should not have an NO level that is likely to last days. Therefore we only reported NO release profiles for the initial 24 h after adding donors, though GSNO and DETA/NO both continued releasing detectable amounts of NO after 24 h under some conditions.
Ten milliliter of PBS with 50 μM of CysNO, SNAP and GSNO and 25 μM of DETA/NO were applied to the NO measurement device in separate experiments. Fig. 4 A to D (blue traces) show their NO release levels. By integrating the area under the release curves, the total NO released in mole can be determined. The total NO released from these donors is recorded in Table 1 . The reported half-lives of each NO donor are also summarized in Table 1 [9] , [22] , [23] and [24] . CysNO showed the most rapid release, while GSNO and DETA/NO generated a gradual and long-term release pattern. However, both the reported SNAP half-life and the NO generation duration measured by the device showed large variations. Fig. 4 E shows each individual repeat for SNAP, which differs greatly from other donors’ consistent and repeatable release profiles.
|
|
|
Fig. 4. The NO release profiles of three different RSNOs and DETA/NO in different buffer conditions are shown. Ten milliliter of 50 μM CysNO, SNAP, GSNO and 25 μM DETA/NO prepared in PBS or DMEM were applied to the CellNO Trap for real-time monitoring, where panels A through D are CysNO, SNAP, GSNO, and DETA/NO, respectively. Under each condition, triplicate experiments were run independently. Data were presented as average (n=3) with error bar representing the standard deviation of the three trials. Panel (E) shows the NO release profile of 50 μM SNAP in 10 ml PBS. Since the release profiles of SNAP showed large variation among replicates, all repeats are presented for readers’ reference. Red marks indicate the highest flux values, red arrows show sharp decrease of the NO flux, green arrow, sharp increase. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) |
| CysNO (50 μM ) | SNAP (50 μM ) | GSNO (50 μM ) | DETA NONOate (25 μM ) | |||
|---|---|---|---|---|---|---|
| Reportedt1/2(in PBS or PSS, h) | 0.023 | 1.15, 4.6, Up to 6 | 159 | 20 | ||
| PBS | w/out CO2 | Total NO (mol) | 7.37E-08 (4.76E-09) | 8.87E-08 (4.87E-08) | 1.09E-07 (6.30E-08) | 9.30E-08 (1.30E-08) |
| Average flux (mol/cm2/s) | 2.49E-12 (1.61E-13) | 4.89E-13 (2.92E-13) | 9.06E-14 (5.21E-14) | 8.11E-14 (1.14E-14) | ||
| Max flux (mol/cm2/s) | 2.12E-11 (1.89E-12) | 1.11E-12 (6.72E-13) | 4.36E-13 (4.80E-13) | 1.19E-13 (1.32E-14) | ||
| Duration (h) | 0.62 (0.024) | 4.04 (2.76) | >24h* | >24h* | ||
| DMEM | w/out CO2 | Total NO (mol) | 1.38E-08 (8.88E-10) | 1.18E-07 (2.96E-09) | 1.36E-07 (2.70E-08) | 8.44E-09 (3.94E-09) |
| Average flux (mol/cm2/s) | 5.76E-13 (3.70E-14) | 6.96E-13 (2.86E-14) | 5.68E-13 (1.25E-13) | 7.22E-15 (3.30E-15) | ||
| Max flu (mol/cm2/s) | 2.65E-12 (1.08E-12) | 3.05E-13 (7.67E-15) | 1.24E-12 (2.22E-13) | 2.55E-14 (1.14E-14) | ||
| Duration (h) | 0.45 (0.056) | 6.42 (0.12) | 22.01 (0.83) | >24h* | ||
| with CO2 | Total NO (mol) | 7.43E-08 (1.14E-09) | 1.10E-07 (2.78E-08) | 1.64E-07 (4.94E-09) | 1.55E-07 (2.30E-08) | |
| Average flux (mol/cm2/s) | 5.07E-13 (1.49E-14) | 7.11E-14 (8.50E-15) | 1.44E-13 (6.42E-15) | 1.32E-13 (1.96E-14) | ||
| Max flux (mol/cm2/s) | 1.73E-12 (6.81E-14) | 1.25E-13 (1.41E-14) | 4.31E-13 (1.59E-14) | 2.33E-13 (3.39E-14) | ||
| Duration (h) | 2.44 (0.46) | >24h* | 4.48 (0.80) | >24h* | ||
The total moles of NO were calculated by integrating the area under the NO release curves; maximum NO flux was directly read from the NO releasing profile; average NO flux refers to the entire releasing duration; t1/2 is the half-life of donors; * duration is longer than 24 h, however we only reported the initial 24 h. Data is presented as the average and (Std. dev.).
The inconsistencies of the NO release from SNAP also casts doubt on how rational it is to use half-life of each NO donor measured in PBS as the reference to predict the NO levels achieved under real biological conditions. To further investigate this in a more biologically relevant scenario, NO release profiles of different NO donors dissolved in DMEM were directly measured. NO donors were dissolved in 10 ml freshly prepared pre-warmed complete DMEM (with 10% FBS and 1% pen/strep) and the CellNO Trap was placed in 37 °C incubator with ambient air for real-time NO monitoring. It should be noted that CO2 was not used in the incubator for this initial work, but because the use of 5% CO2 is a common condition for cell culture and it might potentially affect the solution pH and carbonate concentration, the use of 5% CO2 was considered a separate variable for investigation and will be discussed below. Fig. 4 shows that NO profiles of each NO donor in DMEM without CO2 (red traces). At this specific concentration (50 μM), the NO release profiles of all NO donors differed greatly in PBS from the corresponding release in DMEM (without CO2 ). In DMEM, the total NO that was captured by the sweep gas was 5.4 times and 11.0 times lower for CysNO and DETA/NO, respectively, while SNAP and GSNO both showed that more total NO (1.3 and 1.2 times, respectively) compared with in PBS group within 24 h (Table 1 ). The change of NO flux also diverged; reduced in CysNO, SNAP and DETA/NO but significantly increased in the GSNO samples (Fig. 4 A–D blue traces vs. red traces and quantified in Table 1 ). Those results clearly demonstrated that different buffers do significantly affect NO generation of all four NO donors examined.
When 5% CO2 was applied to the incubator atmosphere, making the conditions more representative of typical cell culture conditions, the NO generation levels were further affected. All the RSNOs showed longer NO release duration in the DMEM with 5% CO2 compared to samples run in DMEM without CO2 (CysNO and SNAP released NO for approximately 2 and 20 h longer, Fig. 4 B, red and green traces, respectively), while DETA/NO released NO in a significant faster fashion (Fig. 4 D, red and green traces). The results might be explained by the lower media pH brought about by CO2 . GSNO and DETA/NO no longer constantly released low level NO for over 24 h but released faster in the initial stage, and after 24 h, the NO level can barely be detected. This might be one of the reasons why the reported GSNO’s half-life is much longer than SNAP’s but in some studies its potency did not follow this trend [9] and [24] . This data clearly shows that simply using an NO donors half-life measured in a simple buffer solution to estimate the level and duration of NO that cells experience is grossly inaccurate.
3.3. Factors that may affect NO release profiles of NO donors
Other factors were also investigated to understand how the level of NO that is present at the cell surface changes when soluble NO donors were used. As previously described, the NO level created by NO donors is determined by both the NO generation and consumption reactions. In first order kinetics model, NO generation can be expressed as follows:
|
|
( 8) |
And in NO auto-oxidation model, the ultimate NO level can be influenced by the consumption described as:
|
|
( 9) |
where:
- [NO] concentration of NO at given time t ,
- t time,
- k1 is the rate constant for the decomposition of the donor,
- [D] is the concentration of the donor at a given time t ,
- eNO is the factor representing moles of NO released per mole of donor,
- [O2 ] is the concentration of oxygen,
- k2 is the rate constant for the oxidation of NO by oxygen,
Any factor that can change generation, consumption or introduce other consumption mechanisms will change the actual NO level obtained. Several known mechanisms can initiate the release of NO from RSNO, including transition metal ions mediated NO releasing, photocleavage, and ascorbate initiated NO release [25] . In addition to direct decomposition, RSNO can undergo transnitrosation reactions with nucleophiles and other thiols without generating any free NO, producing new RSNOs which may affect the NO release profiles as well [25] . Specific factors (presence of transition metals, free thiols, change in pH/CO2 , redox status, and solution volume) were systematically investigated to understand how they affect the ultimate NO level adherent cells experience when these soluble NO donors are used.
3.4. The effect of transition metal ions
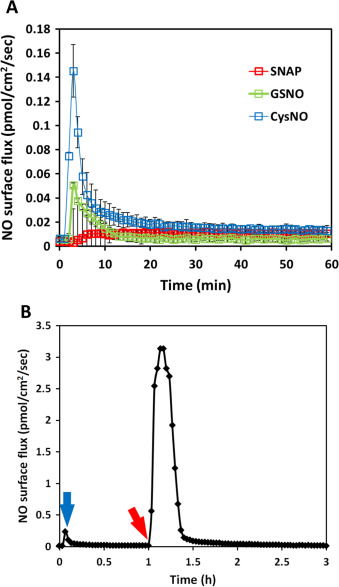
PBS with 10 mM EDTA was used as the NO donor buffer to eliminate the effect of the transition metal ions which are ubiquitous contaminants in buffer salts. Using the same experimental set-up and conditions, SNAP’s NO generation profile under metal ion-free conditions appeared to be significantly slower and more repeatable (red traces in Fig. 5 A). This result suggested that transition metal ions mediated NO releasing is the main release mechanism for SNAP decomposition in PBS, and without transition metal ions, SNAP may be preserved for a much longer time. (After SNAP was placed in 37 °C incubator for 48 h in 10 mM EDTA PBS, 1 mM ascorbic acid was applied to initiate NO release, which rapidly produced a huge amount of NO release, indicating the high stability of SNAP in solutions without transition metal ions (data not shown)). When transition metal ions were removed in CysNO and GSNO solutions, similarly, significant decrease in NO generation was observed in both groups (blue and green traces respectively in Fig. 5 A).
|
|
|
Fig. 5. The NO release profiles of 50 μM of three different RSNOs in 10 ml PBS with 10 mM EDTA. Panel (A) shows the real-time NO release three independent experiments for each RSNO were run and data was presented as the average of the three, error bar representing standard deviation. Panel (B) shows the real-time measured from time 0–1 h of 50 μM CysNO in 10 mM EDTA PBS (blue arrow shows the signal peak); at 1 h, 1 mM ascorbic acid was applied (red arrow), initiating rapid NO release. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) |
After 1 h, the NO generation from both CysNO and GSNO became almost undetectable, but after adding ascorbic acid, NO generation recovered (example shown in Fig. 5 B). The amount of NO generated in the initial 10 min by these three RSNOs in both normal PBS and PBS with 10 mM EDTA are summarized in Table 2 . With transition metal ions RSNOs’ NO generation increased to different degrees, where CysNO generated over 140 times more total NO in the initial 10 min in normal PBS compared with PBS with EDTA while the total NO generation only increased by approximately 50 times and 10 times respectively in SNAP and GSNO. This demonstrates that CysNO is more sensitive to transition metal ions than SNAP and GSNO. Removal of the transition metal ions from RSNOs environment (even CysNO, which is normally considered to be highly reactive), can make RSNOs much more stable, which is consistent with Singh et al.‘s [23] statement that SNAP’s stability can be significantly longer in transition metal ion free solutions. The NO generation from RSNOs in untreated buffers and growth media may be mainly through the transition metal ion mediated mechanism at 37 °C.
| CysNO (50 μM ) | SNAP (50 μM ) | GSNO (50 μM ) | ||
|---|---|---|---|---|
| PBS | First 10min NO release(mol) | 6.64E-08 (4.12E-09) | 4.69E-09 (2.32E-09) | 2.13E-09 (2.07E-09) |
| PBS with EDTA | Total NO (mol) | 1.12E-09 (2.41E-10) | 1.53E-09 (5.97E-10) | 3.53E-11 (7.80E-12) |
| Average flux (mol/cm2/s) | 2.97E-14 (6.75E-15) | 8.88E-15 (2.06E-15) | 5.43E-14 (2.84E-14) | |
| Max flux (mol/cm2/s) | 1.64E-13 (2.42E-14) | 1.46E-14 (3.22E-15) | 8.99E-15 (1.71E-15) | |
| First 10min NO release (mol) | 4.74E-10 (1.07E-10) | 8.24E-11 (2.49E-11) | 2.05E-10 (1.47E-10) | |
| Ratio (with TIs/without TIs) | 140.1 | 56.9 | 10.4 | |
NO moles were calculated by integrating the area under NO releasing curve between 2 specific time points; first 10 min was chosen because in PBS solutions CysNOs NO releasing in PBS and all RSNOs' releasing in 10 mM EDTA PBS became almost undetectable. Data is presented as the average and (Std. dev.).
3.5. The effect of free thiols
The free thiols of GSH and Cys were added to the NO generating solutions to achieve a final concentration of 500 μM to examine their effect on the overall NO levels cells experience. It is noted that high concentration of thiol (mM range) should be avoided because of the potential to change the pH of the buffer. When GSH was directly added to a solution of DETA/NO in PBS, which releases NO in a continuous and near constant rate fashion at 37 °C, a significant NO drop was observed (Fig. 6 A), suggesting the existence of NO-GSH or DETA/NO-GSH interaction. However, after approximately 4 h, the NO level started to return, suggesting this inhibition of NO release was reversible. To further investigate the effect of thiol content on other donor’s NO generation profiles, the same amount of GSH was added to the SNAP PBS solution, but no significant change in the NO release profile was observed (data not shown). In contrast, when the same amount of Cys was added to SNAP, there was a significant reduction in total NO and average flux (see Table 3 ).
|
|
|
Fig. 6. The NO level was changed by introducing free thiols, in panel (A), 10 ml of 100 μM DETA/NO solution was applied to the device for NO flux measurement until the signal reached steady state. The NO flux increased immediately after introducing PBS and mixing as a control (red arrow), then 200 μM GSH was added (blue arrow), causing the level of NO to sharply decrease but it returned to the initial level of NO after approximately 4 h; Panel (B) shows the effect when 10 ml of 50 μM SNAP dissolved in PBS was applied to a solution of 500 μM Cys (n=3). The presences of the free thiol (Cys) made the level of NO produced by SNAP decomposition more reproducible when compared to NO release without the free thiol (see Fig. 4 , Panel E). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) |
| SNAP (50 μM ) | |||
|---|---|---|---|
| PBS | Normal PBS | Total NO (mol) | 8.87E-08 (4.87E-08) |
| Average flux (mol/cm2/s) | 4.89E-13 (2.92E-13) | ||
| Max flux (mol/cm2/s) | 1.11E-12 (6.72E-13) | ||
| Duration (h) | 4.04 (2.76) | ||
| With Cys | Total NO (mol) | 1.26E-07 (2.30E-9) | |
| Average flux (mol/cm2/s) | 3.39E-13 (4.56E-14) | ||
| Max flux (mol/cm2/s) | 5.38E-13 (9.02E-14) | ||
| Duration (h) | 7.54 (0.97) |
NO moles were calculated by integrating the area under NO release curves. Data presented as average (Std.dev.).
Table 3 summarized the NO release parameters observed, clearly showing that instead of greatly changing those numbers, the addition of thiols significantly reduced the variations, making the result more repeatable. Interestingly, in DMEM, the NO generation from SNAP is observed to be more repeatable and the profile increases slowly and decreases slowly, indicating that thiols may be a factor that account for this slowly increasing NO release. Free thiols are important NO reactants in the body. They can elongate the effective NO release and help with NO transportation [26] . They also serve as important regulators of protein nitrosation [27] . Understanding the relationship between NO and thiols can help us understand the underlying mechanisms affecting NO levels present in biological systems.
3.7. The effect of pH and CO2
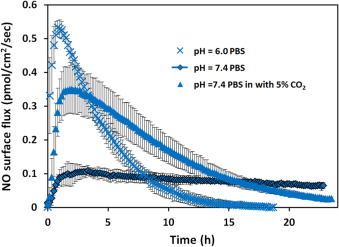
Different tissues have different pHs, which may result in different NO generation profiles from the same NO donor [28] and [29] . Fig. 7 showed the NO release profile of 25 μM DETA/NO in PBS solutions with different pHs. At pH 6 although total NO that cells might experience was only around 1.4 times larger (within 24 h), the maximum NO flux increased around 4.6 times and NO release duration was shortened significantly to around 16 h. Considering the biological effect of NO is determined by the level and duration of NO delivery that specific cells experience, this data indicates specific NO donors may have different potencies in different tissues or cell compartments based on the pH of the target cellular environment.
|
|
|
Fig. 7. The NO release profiles of 25 μM DETA/NO in PBS (pH=7.4 and 6.0) and PBS (pH=7.4 with 5% CO2 ). Three independent experiments under each different condition were run and data was presented as average of the three, error bars representing standard deviation. |
Because blood is a pH buffered system that relies on the balance between CO2 and HCO3- , it is hypothesized that 5% CO2 within the incubator may exert similar effects on the NO level by changing pH of the solution. To test this, PBS buffer was applied to the CellNO Trap and placed into the cell culture incubator with 5% of CO2 . Direct measurement of the pH of the PBS shows that the solution pH decreased to around 6.6 within 90 min (data not shown). Using pH=7.4 PBS buffer to prepare 25 μM DETA/NO with 5% CO2 , demonstrates that CO2 has a significant effect on the NO generation profiles (see Fig. 7 ). One fold increase of total NO and almost 3 times greater maximum flux compared with without CO2 were observed (see Table 4 ). After 24 h, the NO signal was barely detectable. The decomposition of diazeniumdiolates to generate NO has been shown to be pH dependent [30] . Factors that affect the dissociation rates of diazeniumdiolates include pH, concentration and the presence of metal ions [31] and [32] . Protonation is necessary for the decomposition reaction and thus dependent on the pH [33] . The notion that DETA/NO releases NO in a long-lasting fashion might be true only in simple buffer conditions, but not in more complex solutions such as culture medium. The micro-environments of many inflammation situation and tumor masses can achieve relatively low pH value in vivo (even lower than pH=6 [28] ), which is likely to greatly change the effective NO delivery pattern.
| DETA/NO (25 μM ) | |||
|---|---|---|---|
| PBS | pH=7.4 PBS | Total NO (mol) | 9.30E-08 (1.30E-08) |
| Average flux (mol/cm2/s) | 8.11E-14 (1.14E-14) | ||
| Max flux (mol/cm2/s) | 1.19E-13 (1.32E-14) | ||
| Duration (h) | >24h* | ||
| pH=6.0 PBS | Total NO (mol) | 1.31E-07 (1.81E-08) | |
| Average flux (mol/cm2/s) | 1.69E-13 (2.63E-14) | ||
| Max flux (mol/cm2/s) | 5.48E-13 (2.59E-14) | ||
| Duration (h) | 15.86 (1.19) | ||
| pH=7.4 PBS with CO2 | Total NO (mol) | 1.86E-07 (3.76E-08) | |
| Average flux (mol/cm2/s) | 1.47E-13 (2.96E-14) | ||
| Max flux (mol/cm2/s) | 3.62E-13 (7.37E-14) | ||
| Duration (h) | >24h* | ||
NO moles were calculated by integrating the area under the NO release curves. * means that duration is longer than 24 h, however we only examined the initial 24 h. Data is presented as the average and (Std. dev.).
3.7. The effect of redox environment
Oxidative condition of the cellular environment may also influence the ultimate NO levels obtained. It has been reported by many groups that elevated ROS levels suppressed biological potency of NO. The underlying mechanisms is not clearly understood, but some ROSs directly react with NO, decreasing the NO that cells can access and some are through other mediators [34] and [35] . The influence of ROS (such as peroxide and superoxide) to NO level was investigated by real-time monitoring NO from DETA/NO solutions. Different peroxide sources were used (H2 O2 and di-tert-butyl peroxide(DTBP)) to complete this examination. There was no significant decrease in NO level observed when H2 O2 was applied (data not shown) and only very mild NO decrease was observed when up to 1 mM DTBP was applied into 100 μM DETA/NO solution (Fig. 8 ). And NO level returned to normal after approximately 0.5–1 h, indicating only a mild interaction of peroxide and NO.
|
|
|
Fig. 8. The NO level at the surface where cells grow in the CellNO Trap decreased by introducing peroxide. Panel (A) shows that the NO level changed in accordance with the addition of peroxide (blue arrow) compared to the PBS (red arrow); Panels (B) through (D) illustrate three repeats of the experiment (blue curves representing with peroxide, red the control). Ten milliliter of 100 μM DETA/NO solution were applied to the device for NO flux measurement. Once the NO flux reached the steady state, t-butyl peroxide was added into the solution (to the final concentration of 1 mM). All experiments were run at 37 °C. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) |
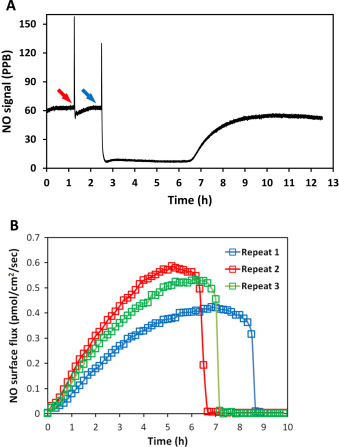
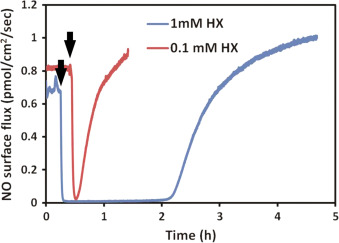
Using the same principle, superoxide was applied to DETA/NO solutions by using hypoxanthine-xanthine oxidase (HX/XO) system [36] . Superoxide generation relies on the availability of the substrate HX. Different concentration (100 μM and 1 mM) of substrate was applied to the system to modulate the amount and duration of superoxide. Data shows that immediately after applying superoxide, the NO signal quickly decreased, indicating that superoxide greatly suppressed the resulting NO level (Fig. 9 ). The NO level recovered gradually to the original level, and the time needed for this recover is correlated to the amount of hypoxanthine added.
|
|
|
Fig. 9. The NO level decreased upon the introduction of superoxide. Hypoxanthine (1 mM in blue and 100 μM in red) was added into 10 ml 100 μM DETA/NO PBS solution pH=7.4 (with 10 mU/ml xanthine oxidase, time points indicated by black arrows). All experiments were run at 37 °C. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) |
3.8. The effect of solution volume
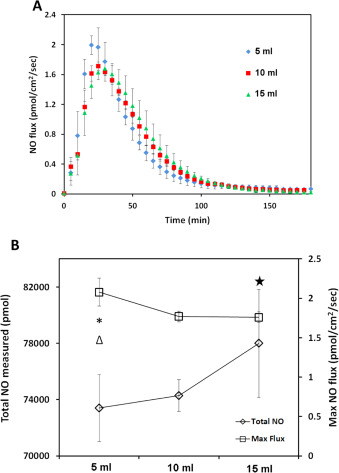
In in'vitro experiments, the quantitative study of NO is normally accomplished by applying media with specific concentration of an NO donor. However, most studies do not precisely specify the final volume of solution in culture wells. Different solution volume may lead to different final NO levels experienced by cell layer at the bottom of the culture vessel, even when the amount of donor added was carefully controlled due to the diffusivity and reactivity of NO through the volume of the liquid medium. To investigate this, 5, 10, 15 ml of 50 μM CysNO DMEM solution was applied to the CellNO Trap and NO levels sustained at the bottom of the culture vessel (which importantly corresponds to the level experienced by an adherent layer of cells) are shown in Fig. 10 A. Keeping all other conditions constant and varying only volume, 5 ml solution generated a peak flux of 2.0 pmol cm−2 min−1 , the highest NO flux (P =0.023, 0.022 compared to 10 ml and 15 ml group respectively shown by * and Δ, P >0.05 between 10 and 15 ml by Tukeys-test), compared to approximately 1.8 and 1.7 pmol cm−2 min−1 for 10 and 15 ml, respectively. There is a decrease in the NO flux level with increasing solution volume, while the total accumulative NO that cell experienced is greater for the larger volumes. The total NO delivered was 73,000, 76,000 and 78,000 pmol for 5, 10 and 15 ml, respectively (see Fig. 10 B (P =0.0370 between 5 and 15 ml groups shown by ⧋, P >0.05 in other groups in Tukeys-test)). Although it is expected that the larger volume of solution would deliver a larger total dose of NO, there is a striking difference in the real-time flux resulting from different volumes of solution.
|
|
|
Fig. 10. Different media volumes illustrate an effect on the NO level experienced by the cultured cells at the bottom of the CellNO Trap. Panel (A), shows the NO release profiles of different volumes of 50 μM CysNO in DMEM with 5% CO2 ; three independent experiments of each volume were run and data was presented as average of the three, error bar representing standard deviation. Panel (B) shows the change of the total NO and NO flux experienced by cells along with the change of the media volume, * and Δ indicate a significant difference of the total NO in between 5 ml and 10 ml, and between 5 ml and 15 ml respectively, P<0.05 by ANOVA and Tukeys test; star indicates a significant difference of the maximum NO flux between 5 ml and 15 ml, P <0.05 by ANOVA and Tukeys test. |
3.9. Different NO donors have different ultimate effect on MOVAS cells
In the previous section, NO release profiles of the four NO donors in real culture conditions (DMEM at 37 °C and 5% CO2 ) were measured. With this information regarding predicted NO fluxes, the inhibition of MOVAS cell proliferation was studied [37] , [38] and [39] .
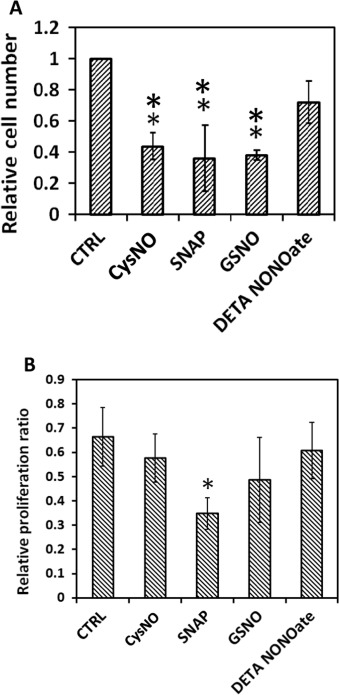
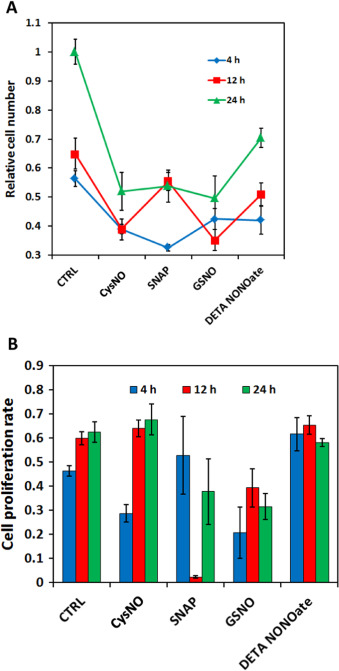
The same four NO donors with the same effective NO concentration (50 μM for RSNOs, and 25 μM for DETA/NO) were used to treat MOVAS cells. After 24 h both cell number and cell proliferation ratio were examined as shown in Fig. 11 . Result shows that DETA/NO treatment had the least potent effect on restricting cell numbers after 24 h. All three RSNOs showed significantly greater inhibitory effect on cell growth and proliferation as indicated by ANOVA and Tukeys test, however, Tukeys test did not suggest any statistic difference existed among the three RSNO groups, which is difficult to explain by the NO real-time profiles obtained using DMEM media with CO2 . Meanwhile cell proliferation assays implicated that after 24 h only SNAP treated cell proliferation ratio differed significantly from the control group (P=0.0565 by ANOVA and P<0.05 by t -test), while cell proliferation ratios of all other groups were slightly smaller but close to the control group (P >0.2 by ANOVA).
|
|
|
Fig. 11. Different NO donors exert different effects on MOVAS cell proliferation. Cell seeding density was 10,000 cell/cm2 and before applying the NO donors, cells were cultured overnight to allow recovery; 24 h after treatment, cells were screened for live-dead assay and proliferation assay (A) Relative living cell number. ** represents P<0.01 by ANOVA and Tukeys-test compared with the control (CTRL). (B) Cell proliferation ratio. * represents P=0.0565 by ANOVA and Tukeys-test but P<0.05 by t -test compared with the CTRL. Data represents the average of the 3 independent repeats and error bar represents standard deviation among. |
The CellNO Trap was used to obtain real-time NO measurements during the actual cell culture to allow correlation the level of NO experienced by the cells with the cell proliferation. Result and quantification analysis were summarized in Fig. 12 A and Table 5 . This clearly shows that, with cells (under actual cell culture conditions) GSNO did not release NO in low and long-lasting fashion as people normal think, but DETA/NO did. Instead GSNO’s release was more similar to CysNO, which released most of its NO in the initial 4 h of the experiment. Compared with the media only condition, the average and maximum flux of all RSNOs increased significantly (around 2, 4, and 9 times more for CysNO, SNAP and GSNO respectively), though the total accumulated NO experienced by cells did not change much (a little over 1 fold for CysNO and only 50% more and 20% less for SNAP and GSNO). And both NO flux and total NO diffusing to and through the cell layer greatly decreased in the DETA/NO treated group (only 30% and 15% compared with media only conditions).
|
|
|
Fig. 12. The real-time measurement of the NO levels that cells experienced when in panel (A) they were cultured in the collagen surface-treated CellNO Trap and treated with 10 ml of media with 50 μM CysNO, SNAP, GSNO or 25 μM DETA/NO; and panel(B) shows cells cultured on the polydopamine surface-treated CellNO Trap when treated with 10 ml of media containing 50 μM CysNO, SNAP, GSNO or 25 μM DETA/NO. Under each condition, triplicate experiments were run independently. Data was presented as the average value of the triplicate; error bar represents the standard deviation of the three values. |
| CysNO (50 μM ) | SNAP (50 μM ) | GSNO (50 μM ) | DETA/NO (25 μM ) | |||
|---|---|---|---|---|---|---|
| Complete media with cells | Collagen I coating with cells | Total NO (mol) | 1.67E-07 (5.17E-08) | 1.71E-07 (2.39E-08) | 1.32E-07 (2.15E-08) | 2.54E-08 (4.21E-09) |
| Average flux (mol/cm2/s) | 1.07E-12 (2.56E-13) | 3.77E-13 (8.33E-14) | 4.70E-13 (3.35E-14) | 3.21E-14 (1.05E-14) | ||
| Max flux (mol/cm2/s) | 4.96E-12 (1.18E-12) | 5.92E-13 (7.42E-14) | 1.80E-12 (3.58E-13) | 7.38E-14 (3.40E-15) | ||
| Duration (h) | 3.15 (0.98) | 9.46 (2.55) | 4.44 (0.43) | 14.60 (3.12) | ||
| Dopamine coating with cells | Total NO (mol) | 1.30E-07 (3.15E-08) | 1.87E-07 (4.92E-08) | 1.31E-07 (2.60E-08) | 9.30E-08 (3.20E-08) | |
| Average flux (mol/cm2/s) | 6.86E-13 (4.68E-13) | 4.61E-13 (9.00E-14) | 5.95E-13 (1.05E-13) | 1.38E-13 (7.42E-14) | ||
| Max flux (mol/cm2/s) | 2.67E-12 (5.46E-13) | 8.40E-13 (2.94E-14) | 1.24E-12 (3.85E-13) | 2.48E-13 (9.03E-14) | ||
| Duration (h) | 2.76 (0.19) | 5.33 (2.20) | 4.06 (0.64) | 13.15 (1.06) | ||
Cells were cultured in either a collagen I top-coated or a dopamine-gelatin treated CellNO Trap device, respectively. NO donors were dissolved in media to twice the desired final concentration in equal volume media to original cell culture media and applied to the cultured cells. Data is presented as the average and (Std. dev.).
This data clearly demonstrates that during actual cell culture experiment, the NO donors may release NO in a very different manner than predicted from the release profile obtained from the controlled experimental conditions. Simply knowing that different NO donors have different NO releasing profiles is not enough to explain some of their biological outcomes. It is also important to remember that a donor’s NO delivery behavior is highly changeable, and may result in a totally different profile when experimental conditions are changed slightly. For example, CysNO and GSNO are normally considered to have very different stability, but actually both of them released NO rapidly in the initial stages of the in vitro experiment with a very high flux (>1 pmol/cm2 /s, Fig. 12 A), and no NO was detected by the end of the 24 h treatment. This result explains the data shown in Fig. 11 as to why the cell number and cell proliferation ratio in CysNO and GSNO group were very similar, and DETA/NOs least potency may be resulting from the very low NO level generated during the actual cell culture experiment. SNAP has an intermediate NO level in real cell culture conditions (around 0.4 pmol/cm2 /s) and lasted a significant longer time compared with other RSNO (persisted up to 12 h duration), this might be one of the reasons why after 24 h the smallest cell proliferation ratio was observed in SNAP group. But overall, the SNAP treated group did not show significantly smaller cell number after 24 h compared with the CysNO and GSNO groups. This observed inconsistency between cell number and cell proliferation ratio may result from the fluctuation of cell proliferation ratios within the 24 h experiment time because of the changing of the NO level. To better understand NO’s effect on MOVAS, instead of doing a single end-point examination on cells, cells need to be examined at multiple intermediate time points during the course of the 24 h experiment.
3.10. NO profile and cell proliferation at different time points
Cell number and cell proliferation ratios were examined at different time points to assess the effect of the selection of time points in end-point testing. According to the NO profile data Fig. 12 (A), CysNO and GSNO’s NO release lasted for around 3–4 h, and at 11–12 h SNAP finished releasing NO, this lead to the examination of cells at 4 h and 12 h and 24 h.
Cell number data clearly showed that even though at 24 h cell numbers in three RSNO groups were similar, the change of cell number/proliferation ratio was very different at the intermediate time points (Fig. 13 A). The increase of cell number in CysNO and GSNO groups was mainly after 12 h and between 4 and 8 h the increase is low, however, SNAP showed the inverse pattern, where the increase was mainly in the first 12 h. DETA/NO has relatively stable with a slow increase of cell number. And these results are also consistent with the cell proliferation ratio data (Fig. 13 B), where in CysNO and GSNO treatments the smallest proliferation ratios were observed at 4 h, and in the SNAP treatment, the lowest value was at 12 h. DETA/NO showed a consistently high cell proliferation ratios, but the resultant cell numbers were still smaller than the controls (Fig. 13 A). By using the CellNO Trap, it is demonstrated that an NO flux of around 0.4–1 pmol/cm2 /s on average showed good inhibition of MOVAS cell proliferation. A constantly high flux (around >1.5 pmol lasting for over 2 h) may kill the cells (data not shown). It has now been clearly illustrated that the actual NO status experienced during the culture experiment is critical, not the empirical values determined or measured under other conditions (for example, it is not recommended to use half-lives of NO donors’ measured in PBS or complete media even with 5% CO2 to explain observed biological data). The results presented here directly show the significance of tracking the actual NO level generated by NO donors in biological experiments throughout the duration of the culturing experiments.
|
|
|
Fig. 13. Different NO donors inhibited MOVAS cell proliferation to different degrees at different time points. Panel (A) shows the relative cell numbers normalized to cell number of control groups at 24 h and panel (B) shows the MOVAS’s cell proliferation ratio under different NO donor treatments at different time points. Cell seeding density was 10,000 cell/cm2 and before applying NO donors cells were cultured overnight to allow recovery; 4, 12 and 24 h after treatment, cells were stained for life-death assay and proliferation assay. Data represents the average of 3 independent repeats and error bar represents sample standard deviation. |
Though NO’s biological roles became much clearer during the past several decades, many contradictions and enigmas still exist. One problem is that NO’s effect on cells in cell culture is hard to be separated from the behavior of soluble NO donors. One systematic study on biological effect of different RSNO donors was reported by Mathews et al.[9] . Considering the general idea that RSNO’s effect was through NO released from the cleavage of S-NO bond, they designed gas-purging experiment which showed that removing NO had negligible influence on smooth muscle relaxation, suggesting that at least other activation pathways to soluble guanylate cyclase (sGC) exist. They also showed that the correlation between RSNO decomposition first-order half-life and the degree of the smooth muscle relaxation to be low. However, the half-life values they used were measured within 0.5 mM physiologic saline solution, while the biological assays were in more complicated systems. Wink et al. [10] showed NOs cyto-protective effect on hydrogen peroxide stressed V79 cells and no protective effect on superoxide stress, even after superoxide dismutase (SOD) was added. However, in the same paper they also showed different NO donors of the same working concentration had very different potencies in counteracting peroxide mediated cytotoxicity, showing that choosing the proper NO donor is critical. Using a system with both HX/XO and RSNO showed more complicated behavior according to Trujillo et al. [40] , where peroxynitrite might form and stress cells when simultaneously using these two chemicals in the same system. Sodium nitroprusside (SNP) was shown to be cytotoxic because of the synergetic effect of peroxide and the release of other ions such as CN− and iron complex [41] , and 3-morpholinosydnonimine (SIN-1) may facilitate the accumulation of H2 O2 for cell cancer cytotoxicity [42] and [43] . So tracking the real-time NO status is critical in helping researchers evaluate the net effect of NO in complicated systems. Instead of using new NO donors, some groups developed NO delivery systems that can directly deliver only NO into the biological systems [44] , [45] and [46] . Though problems still exist such as the controllability, consumption of NO through the biological system, biocompatibility and cytotoxic effect of NO’s oxidative products, the use of NO releasing polymer and device is still promising approaches to understand the effects NO exerts on cellular systems.
A huge number of potential biological species can react with NO, including ROS [34] and [35] , thiols [47] , lipid [48] , DNA [49] and [50] , and proteins [51] , [52] and [53] , which makes the actual NO level present in cellular environments hard to predict. For example, the effect of free thiol to NO is complicated. On one hand, thiols may react with NO forming new RSNOs, which may increase the effective half-life of NO but decrease the effective NO level. On the other hand, free thiol can facilitate RSNO decomposition resulting in a more rapid NO generation [54] . And these processes are very likely to be pH dependent. For RSNOs, lower pH potentially generates more HNO2 and NO+ , which are strong nitrosating agents, preventing NO generation [55] , while for diazeniumdiolates NO releasing is through the protonation, which favors lower pH [33] . In biological system, cases can be more complicated because of different environments such as ion strength [31] , making final NO level difficult to predict. To investigate the net biological effect of NO, the CellNO Trap device directly measures NO that cells experience, helping us understand the biological function of NO.
4. Conclusions
The NO generation profiles of different NO donors (CysNO, SNAP, GSNO and DETA/NO) at physiological relevant levels in different buffer conditions were examined, proving that NO generation profiles of all NO donors can be very dynamically affected by their specific environments by using the CellNO Trap, a two-chamber real-time NO measurement device developed by our lab previously. This information allows the direct correlation of NO level (not NO donor concentration or NOS level) with the observed biological response. MOVAS cells was directly cultured in the device and treated with different NO donors, while at the same time the actual NO experienced by cells during the whole culture was tracked. The result showed that NO experienced by cells cannot be simply predict or estimated by the NO donor half-life or from a measured value obtained in PBS. The different effects of NO shown by different NO donors on inhibiting MOVAS proliferation was studies by using CellNO Trap, highlighting the importance of choosing correct examination time points and tracking NO duration of release. To precisely study NO’s biological effect, it is highly recommended to monitor and report real-time NO level of each specific experiment to eliminate the reporting of complicating circumstances that lead to inaccurate results.
References
- [1] C. Bogdan; Nitric oxide and the immune response; Nat. Immunol., 2 (10) (2001), pp. 907–916
- [2] E.M. Schuman, D.V. Madison; Nitric oxide and synaptic function; Annu. Rev. Neurosci., 17 (1994), pp. 153–183
- [3] L.J. Ignarro; Nitric oxide: a unique endogenous signaling molecule in vascular biology (Nobel lecture); Angew. Chem. Int. Ed., 38 (13–14) (1999), pp. 1882–1892
- [4] M. Cooke, P. John, M. Dzau, J. Victor; Nitric oxide synthase: role in the genesis of vascular disease; Annu. Rev. Med., 48 (1) (1997), pp. 489–509
- [5] E. Langford, R. Wainwright, J. Martin; Platelet activation in acute myocardial infarction and unstable angina is inhibited by nitric oxide donors; Arter. Thromb. Vasc. Biol., 16 (1) (1996), pp. 51–55
- [6] I. Megson, et al.; Prolonged effect of a novel S-nitrosated glyco-amino acid in endothelium-denuded rat femoral arteries: potential as a slow release nitric oxide donor drug; Br. J. Pharmacol., 122 (8) (1997), pp. 1617–1624
- [7] I. Megson, et al.; N-substituted analogues of S-nitroso-N-acetyl-D, L-penicillamine: chemical stability and prolonged nitric oxide mediated vasodilatation in isolated rat femoral arteries; Br. J. Pharmacol., 126 (3) (1999), pp. 639–648
- [8] A.B. Levine, D. Punihaole, T.B. Levine; Characterization of the role of nitric oxide and its clinical applications; Cardiology, 122 (1) (2012), pp. 55–68
- [9] W.R. Mathews, S.W. Kerr; Biological activity of S-nitrosothiols: the role of nitric oxide; J. Pharmacol. Exp. Ther., 267 (3) (1993), pp. 1529–1537
- [10] D.A. Wink, et al.; The effect of various nitric oxide-donor agents on hydrogen peroxide-mediated toxicity: a direct correlation between nitric oxide formation and protection; Arch. Biochem. Biophys., 331 (2) (1996), pp. 241–248
- [11] K.B. Sandau, J. Fandrey, B. Brüne; Accumulation of HIF-1α under the influence of nitric oxide; Blood, 97 (4) (2001), pp. 1009–1015
- [12] D.A. Wink, et al.; Nitric oxide and some nitric oxide donor compounds enhance the cytotoxicity of cisplatin; Nitric Oxide, 1 (1) (1997), pp. 88–94
- [13] J.R. Lancaster; A tutorial on the diffusibility and reactivity of free nitric oxide; Nitric Oxide, 1 (1) (1997), pp. 18–30
- [14] B.G. Hill, et al.; What part of NO don’t you understand? Some answers to the cardinal questions in nitric oxide biology; J. Biol. Chem., 285 (26) (2010), pp. 19699–19704
- [15] B.Y. Owusu, R. Stapley, R.P. Patel; Nitric oxide formation versus scavenging: the red blood cell balancing act; J. Physiol., 590 (20) (2012), pp. 4993–5000
- [16] W. He, M.C. Frost; CellNO Trap: novel device for quantitative, real-time, direct measurement of nitric oxide from cultured RAW 267.4 macrophages; Redox Biol.,, 8 (2016), pp. 383–397
- [17] B.K. Yang, et al.; Methodologies for the sensitive and specific measurement of S-nitrosothiols, iron-nitrosyls, and nitrite in biological samples; Free Radic. Res., 37 (1) (2003), pp. 1–10
- [18] T.W. Hart; Some observations concerning the S-nitroso and S-phenylsulphonyl derivatives of L-cysteine and glutathione; Tetrahedron Lett., 26 (16) (1985), pp. 2013–2016
- [19] K. Schmidt, et al.; Release of nitric oxide from donors with known half-life: a mathematical model for calculating nitric oxide concentrations in aerobic solutions; Naunyn-Schmiedebergs Arch. Pharmacol., 355 (4) (1997), pp. 457–462
- [20] G.E. Gierke, M. Nielsen, M.C. Frost; S-Nitroso-N-acetyl-d -penicillamine covalently linked to polydimethylsiloxane (SNAP–PDMS) for use as a controlled photoinitiated nitric oxide release polymer ; Sci. Technol. Adv. Mater., 12 (5) (2011), p. 055007
- [21] J.S. Beckman, W.H. Koppenol; Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly; Am. J. Physiol. – Cell Physiol., 271 (5) (1996), pp. C1424–C1437
- [22] C.M. Maragos, et al.; Complexes of ·NO with nucleophiles as agents for the controlled biological release of nitric oxide. Vasorelaxant effects; J. Med. Chem., 34 (11) (1991), pp. 3242–3247
- [23] R.J. Singh, et al.; Mechanism of nitric oxide release from S-nitrosothiols; J. Biol. Chem., 271 (31) (1996), pp. 18596–18603
- [24] D.L. Mooradian, T.C. Hutsell, L.K. Keefer; Nitric oxide (NO) donor molecules: effect of NO release rate on vascular smooth muscle cell proliferation in vitro; J. Cardiovasc. Pharmacol., 25 (4) (1995), pp. 674–678
- [25] D.L.H. Williams; The chemistry of S-nitrosothiols; Acc. Chem. Res., 32 (10) (1999), pp. 869–876
- [26] J.S. Stamler, et al.; S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds; Proc. Natl. Acad. Sci. USA, 89 (1) (1992), pp. 444–448
- [27] V.G. Kharitonov, A.R. Sundquist, V.S. Sharma; Kinetics of nitrosation of thiols by nitric oxide in the presence of oxygen; J. Biol. Chem., 270 (47) (1995), pp. 28158–28164
- [28] I.F. Tannock, D. Rotin; Acid pH in tumors and its potential for therapeutic exploitation; Cancer Res., 49 (16) (1989), pp. 4373–4384
- [29] J. Wike-Hooley, J. Haveman, H. Reinhold; The relevance of tumour pH to the treatment of malignant disease; Radiother. Oncol., 2 (4) (1984), pp. 343–366
- [30] T. Hansen, A. Croisy, L. Keefer; N-nitrosation of secondary amines by nitric oxide via the 'Drago complex'; IARC Sci. Publ., 41 (1981), pp. 21–29
- [31] J.A. Hrabie, L.K. Keefer; Chemistry of the nitric oxide-releasing diazeniumdiolate (“nitrosohydroxylamine”) functional group and its oxygen-substituted derivatives; Chem. Rev., 102 (4) (2002), pp. 1135–1154
- [32] L.K. Keefer; Progress toward clinical application of the nitric oxide-releasing diazeniumdiolates 1; Annu. Rev. Pharmacol. Toxicol., 43 (1) (2003), pp. 585–607
- [33] K.M. Davies, et al.; Chemistry of the diazeniumdiolates. 2. Kinetics and mechanism of dissociation to nitric oxide in aqueous solution; J. Am. Chem. Soc., 123 (23) (2001), pp. 5473–5481
- [34] D.A. Wink, et al.; Nitric oxide protects against cellular damage and cytotoxicity from reactive oxygen species; Proc. Natl. Acad. Sci., 90 (21) (1993), pp. 9813–9817
- [35] D.A. Wink, et al.; Nitric oxide (NO) protects against cellular damage by reactive oxygen species; Toxicol. Lett., 82 (1995), pp. 221–226
- [36] T. Sawa, T. Akaike, H. Maeda; Tyrosine nitration by peroxynitrite formed from nitric oxide and superoxide generated by xanthine oxidase; J. Biol. Chem., 275 (42) (2000), pp. 32467–32474
- [37] F.C. Tanner, et al.; Nitric oxide modulates expression of cell cycle regulatory proteins A cytostatic strategy for inhibition of human vascular smooth muscle cell proliferation; Circulation, 101 (16) (2000), pp. 1982–1989
- [38] U.C. Garg, A. Hassid; Nitric oxide-generating vasodilators and 8-bromo-cyclic guanosine monophosphate inhibit mitogenesis and proliferation of cultured rat vascular smooth muscle cells; J. Clin. Investig., 83 (5) (1989), p. 1774
- [39] K.-I. Kariya, et al.; Antiproliferative action of cyclic GMP-elevating vasodilators in cultured rabbit aortic smooth muscle cells; Atherosclerosis, 80 (2) (1989), pp. 143–147
- [40] M. Trujillo, et al.; Xanthine Oxidase-mediated decomposition of S-Nitrosothiols; J. Biol. Chem., 273 (14) (1998), pp. 7828–7834
- [41] J.M. Campbell, et al.; The interaction of sodium nitroprusside with peripheral white blood cells in vitro: a rationale for cyanide release in vivo; Biochim. Biophys. Acta – Gen. Subj., 1156 (3) (1993), pp. 327–333
- [42] D. Gergeľ, et al.; Increased cytotoxicity of 3-morpholinosydnonimine to HepG2 cells in the presence of superoxide dismutase role of hydrogen peroxide and iron; J. Biol. Chem., 270 (36) (1995), pp. 20922–20929
- [43] R. Farias-Eisner, et al.; The chemistry and tumoricidal activity of nitric oxide/hydrogen peroxide and the implications to cell resistance/susceptibility; J. Biol. Chem., 271 (11) (1996), pp. 6144–6151
- [44] C. Wang, W.M. Deen; Nitric oxide delivery system for cell culture studies; Ann. Biomed. Eng., 31 (1) (2003), pp. 65–79
- [45] M.A. Starrett, et al.; Wireless platform for controlled nitric oxide releasing optical fibers for mediating biological response to implanted devices; Nitric Oxide, 27 (4) (2012), pp. 228–234
- [46] G.E. Romanowicz, et al.; Novel device for continuous spatial control and temporal delivery of nitric oxide for in vitro cell culture ; Redox Biol., 1 (1) (2013), pp. 332–339
- [47] R. Radi, et al.; Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide; J. Biol. Chem., 266 (7) (1991), pp. 4244–4250
- [48] R. Radi, et al.; Peroxynitrite-induced membrane lipid peroxidation: the cytotoxic potential of superoxide and nitric oxide; Arch. Biochem. Biophys., 288 (2) (1991), pp. 481–487
- [49] D.A. Wink, K.S. Kasprzak; DNA deaminating ability and genotoxicity of nitric oxide and its progenitors; Science, 254 (5034) (1991), p. 1001
- [50] T. Nguyen, et al.; DNA damage and mutation in human cells exposed to nitric oxide in vitro; Proc. Natl. Acad. Sci. USA, 89 (7) (1992), pp. 3030–3034
- [51] J.S. Stamler; Redox signaling: nitrosylation and related target interactions of nitric oxide; Cell, 78 (6) (1994), pp. 931–936
- [52] F.J. Schopfer, P.R. Baker, B.A. Freeman; NO-dependent protein nitration: a cell signaling event or an oxidative inflammatory response?; Trends Biochem. Sci., 28 (12) (2003), pp. 646–654
- [53] R. Radi; Nitric oxide, oxidants, and protein tyrosine nitration; Proc. Natl. Acad. Sci. USA, 101 (12) (2004), pp. 4003–4008
- [54] T.-M. Hu, T.-C. Chou; The kinetics of thiol-mediated decomposition of S-nitrosothiols; AAPS J., 8 (3) (2006), pp. E485–E492
- [55] M.K., Morakinyo, S-Nitrosothiols: Formation, Decomposition, Reactivity and Possible Physiological Effects. 2010.
Document information
Published on 20/10/16
Licence: Other
Share this document
claim authorship
Are you one of the authors of this document?