Abstract
Background
Dilated Cardiomyopathy (DCM) is one of the most commonly encountered heart diseases reported globally. It is characterized by enlarged ventricles with impaired systolic and diastolic functions. Mutations in LMNA gene are one of the causative factors to precipitate the disease. However, association of SNPs of LMNA with DCM in particular has not been well documented.
Method
Here we present a limited and restricted case study of patients from south eastern part of India afflicted with idiopathic DCM and conduction defects. By using next generation sequencing we have sequenced the exons of LMNA gene from genomic DNA isolated from patients.
Result
We have identified the linkage of 8 different LMNA SNPs with idiopathic DCM viz. rs121117552, rs538089, rs505058, rs4641, rs646840, rs534807, rs80356803 and rs7339. These SNPs are scattered throughout the gene with prevalence for the region encoding the central rod domain of lamin A/C.
Conclusion
Most of these SNPs in LMNA were previously reported to be involved in various disorders other than DCM. We conclude that, variation in LMNA is one of the major underlying genetic causes for the pathogenesis of DCM, as observed in few Indian populations.
Keyword
Dilated Cardiomyopathy;LMNA;Single Nucleotide Polymorphism;Echocardiography
1. Introduction
Dilated Cardiomyopathy (DCM) is a disease of the heart muscle which is characterized by ventricular dilation and reduced myocardial contractility thereby resulting into impaired systolic and diastolic function [1]. Clinical symptoms include heart failure, thromboembolism and sudden cardiac death. DCM is the most common among the five commonly characterized cardiomyopathies. It has an estimated prevalence of 1/2500 among different populations [2] ; [3]. The pattern of the disease inheritance is mostly autosomal dominant [4]. However, genetic screening shows only 30–35% familial DCM follow the Mendelian mode of disease inheritance [5] while the remaining idiopathic origin DCM follows a complex multivariant origin. Extensive epidemiological studies are still limited to US, European, and Australian populations and also to some Asian countries like Japan, China and Korea where a total of 165 LMNA mutations have been reported (http://www.umd.be/LMNA/) [6]; [7]; [8]; [9]; [10]; [11]; [12] ; [13].
More than 40 genes have been reported so far to be associated with the pathogenesis of DCM which is a heterogeneous disease [14]. 6% of all DCM cases are caused by mutations in lamin A/C gene (LMNA). Fatkin et al. in 1999 [15] first showed the involvement of LMNA mutations with DCM and conduction diseases. All these studies suggest that the LMNA related DCM patients portend a high risk of sudden cardiac death.
LMNA consists of 12 exons and encodes two splice variants lamin A and C which maps in the long arm of chromosome 1 (1q21.2–q21.3) [16]. Mutations in LMNA are known to cause a wide spectrum of diseases other than DCM, in a tissue specific manner collectively termed as laminopathies namely Lipodystrophy, Limb–girdle muscular dystrophy, Emery–Dreifuss muscular dystrophy and many more [17] ; [18]. Most LMNA mutations causing striated muscle disorder are missense mutations distributed throughout all the exons of the gene. Along with mutations in LMNA, various Single Nucleotide Polymorphisms (SNPs) in LMNA were reported to be associated with different disorders other than laminopathies. A total of 40 LMNA SNPs are reported in Leiden Open Variation Database (www.dmd.nl/lmna_seqvar.html). Out of 40, 30 are silent mutations and the rest are missense mutations [19]. A frequently occurring LMNA SNP, rs4641 at exon 10 was found to be associated with various disorders such as adipose tissue metabolism and obesity related phenotypes [20]; [21] ; [22]. In spite of having such a high prevalence rate, still LMNA related DCM patients suffer from poor prognosis [11] ; [23], high risk of sudden cardiac death and life threatening arrhythmias. The underlying cause of DCM due to LMNA mutations is still largely unknown and it lacks proper genotype–phenotype correlation. Therefore, the severity of LMNA mutations or variations in DCM patients calls for the genetic testing of LMNA in patients for early prognosis and to clinically manage complications of the disease on a wider population.
Increasing number of patients in West Bengal, India is diagnosed with DCM each year which is a serious health concern. Patients come to the clinic complaining of respiratory distress, cough and chest pain, edema of distal extremities, palpitations and syncopal or presyncopal attack. A combination of investigations according to the recommendations of American Heart Association (AHA) [2] and World Health Organization (WHO) [24] form the major diagnostic approaches for DCM patients. We have specifically focused into such a tertiary care center at Kolkata-N.R.S. Medical College and Hospital which receives a number of patients from Kolkata and its surrounding districts afflicted with DCM. We have confined our studies on IDCM. We report for the very first time the association of LMNA SNPs with IDCM patients of eastern zone of India. Through genetic analysis we have revealed the association of 8 different LMNA SNPs with IDCM patients. Among these 8 SNPs, SNPs rs538089, rs505058, and rs4641 were previously reported to be associated with DCM in French population [25]. The rests of the SNPs rs121117552, rs646840, rs534807, rs80356803, and rs7339 were hitherto reported for other diseases but not DCM.
2. Materials and methods
2.1. Clinical assessment and screening of subjects for DCM
The clinical investigation and management of DCM started with the acquisition of patients history on admission. Following the history of the patients the physicians would diagnose for DCM and screen them, following the recommendations of AHA [2] and WHO guidelines [24]. The investigations included Chest X-ray, ECG and echocardiography and coronary angiography (if needed). Echocardiography is still regarded as the gold standard for diagnosis. Written informed consent was obtained in accordance with the study protocol approved by the local ethical committee. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki. A cohort of 10 unrelated patients with diagnosed IDCM and suitable 12 control individuals were selected for our study from N.R.S. Medical College and Hospital, Kolkata, India.
2.2. Isolation of genomic DNA from peripheral blood samples
4–5 ml of blood was drawn from the antecubital vein and transported in ice from the hospital to the laboratory in a 6 ml sterile EDTA containing vial. The blood was then transferred into the 15 ml polypropylene conical centrifuge tube and the volume was adjusted to 15 ml by adding RBC lysis buffer (150 mM NH4Cl, 1 mM NaHCO3) followed by incubation at room temperature for 15 min. The cells were pelleted at 3000 rpm in a clinical centrifuge. This step was repeated 3–4 times until WBC was found. The supernatant was carefully decanted and 3 ml of nucleic acid lysis buffer (10 mM TRIS pH 8, 400 mM NaCl, 2 mM Na2EDTA, SDS 0.5%) was added followed by the addition of 100 μl proteinase K (10 mg/ml) and vortexed. The sample was then incubated at 56 °C for 1–2 h. Then equal volume of water saturated phenol was added and mixed. The mixture was centrifuged at 12,000 rpm for 12 min and this step was repeated 2–3 times followed by the addition of equal volume of chloroform. It was centrifuged at 12,000 rpm for 12 min and the supernatant was transferred to a fresh tube to which 1/10 volume of 10 M ammonium acetate and 2.5 times ice cold 100% ethanol was added and mixed gently until the precipitate formed. It was then centrifuged at 12,000 rpm for 12 min and the supernatant discarded. The pellet that was formed was washed with 1 ml ice-cold 70% ethanol. After centrifugation the ethanol was carefully aspirated as not to dislodge the pellet which was air dried and subsequently dissolved in 300–500 μl of TE buffer. This constituted the genomic DNA.
2.3. Genetic testing
Genomic DNA isolated from peripheral lymphocytes of subjects was used as a template for genetic testing of the LMNA gene. The LMNA gene was amplified using 29 sets of primers which cover the entire coding region of the LMNA ( Table 1); 12 coding exons as well as the immediate intronic regions. Sequencing was performed in Ion Personal Genome Machine® (PGM™) System using Ion PGM™ Sequencing 200 Kit v2 (following the manufactures protocol). The SNPs obtained from next generation sequencing were further validated by Sanger sequencing. The 12 sets of primers reported in Perrot et al. [9], were used for sequencing the desired exons of LMNA by Sanger sequencing.
| Primer ID | Start position | End position | Amplicon Id | Start position | End position |
|---|---|---|---|---|---|
| (Without the common tail) | |||||
| LMNA_PP1_1F | 156084537 | 156084556 | LMNA_PP1_1 | 156084557 | 156084748 |
| LMNA_PP1_230R | 156084749 | 156084767 | |||
| LMNA_PP1_162F | 156084698 | 156084716 | LMNA_PP1_162 | 156084717 | 156084906 |
| LMNA_PP1_390R | 156084907 | 156084926 | |||
| LMNA_PP1_346F | 156084882 | 156084900 | LMNA_PP1_346 | 156084901 | 156085090 |
| LMNA_PP1_574R | 156085091 | 156085110 | |||
| LMNA_PP2_1F | 156100366 | 156100394 | LMNA_PP2_1 | 156100395 | 156100550 |
| LMNA_PP2_202R | 156100551 | 156100567 | |||
| LMNA_PP2_52F | 156100417 | 156100439 | LMNA_PP2_52 | 156100440 | 156100600 |
| LMNA_PP2_267R | 156100601 | 156100633 | |||
| LMNA_PP3_1F | 156104135 | 156104160 | LMNA_PP3_1 | 156104161 | 156104365 |
| LMNA_PP3_54F | 156104187 | 156104208 | LMNA_PP3_54 | 156104209 | 156104365 |
| LMNA_PP3_250R | 156104366 | 156104385 | |||
| LMNA_PP4_1F | 156104537 | 156104559 | LMNA_PP4_1 | 156104560 | 156104723 |
| LMNA_PP4_210R | 156104724 | 156104751 | |||
| LMNA_PP4_122F | 156104659 | 156104679 | LMNA_PP4_122 | 156104680 | 156104852 |
| LMNA_PP4_334R | 156104853 | 156104872 | |||
| LMNA_PP5_1F | 156104895 | 156104916 | LMNA_PP5_1 | 156104917 | 156105093 |
| LMNA_PP5_220R | 156105094 | 156105115 | |||
| LMNA_PP5_183F | 156105074 | 156105094 | LMNA_PP5_183 | 156105095 | 156105304 |
| LMNA_PP5_230F | 156105123 | 156105146 | LMNA_PP5_230 | 156105147 | 156105304 |
| LMNA_PP5_432R | 156105305 | 156105326 | |||
| LMNA_PP6_1F | 156105515 | 156105540 | LMNA_PP6_1 | 156105541 | 156105739 |
| LMNA_PP6_242R | 156105740 | 156105757 | |||
| LMNA_PP6_204F | 156105715 | 156105738 | LMNA_PP6_204 | 156105739 | 156105958 |
| LMNA_PP6_466R | 156105959 | 156105981 | |||
| LMNA_PP7_1F | 156105981 | 156106000 | LMNA_PP7_1 | 156106001 | 156106170 |
| LMNA_PP7_210R | 156106171 | 156106190 | |||
| LMNA_PP7_81F | 156106052 | 156106081 | LMNA_PP7_81 | 156106082 | 156106251 |
| LMNA_PP7_291R | 156106252 | 156106271 | |||
| LMNA_PP8_1F | 156106608 | 156106626 | LMNA_PP8_1 | 156106627 | 156106797 |
| LMNA_PP8_208R | 156106798 | 156106815 | |||
| LMNA_PP8_82F | 156106688 | 156106709 | LMNA_PP8_82 | 156106710 | 156106871 |
| LMNA_PP8_286R | 156106872 | 156106893 | |||
| LMNA_PP9_1F | 156106868 | 156106887 | LMNA_PP9_1 | 156106888 | 156107040 |
| LMNA_PP9_192R | 156107041 | 156107059 | |||
| LMNA_PP10_1F | 156107323 | 156107342 | LMNA_PP10_1 | 156107343 | 156107531 |
| LMNA_PP10_228R | 156107532 | 156107550 | |||
| LMNA_PP10_147F | 156107463 | 156107483 | LMNA_PP10_147 | 156107484 | 156107652 |
| LMNA_PP10_350R | 156107653 | 156107672 | |||
| LMNA_PP10_230F | 156107552 | 156107570 | LMNA_PP10_230 | 156107571 | 156107756 |
| LMNA_PP10_459R | 156107757 | 156107788 | |||
| LMNA_PP11_1F | 156108192 | 156108209 | LMNA_PP11_1 | 156108210 | 156108396 |
| LMNA_PP11_223R | 156108397 | 156108415 | |||
| LMNA_PP11_176F | 156108367 | 156108384 | LMNA_PP11_176 | 156108385 | 156108547 |
| LMNA_PP11_381R | 156108548 | 156108575 | |||
| LMNA_PP11_241F | 156108432 | 156108451 | LMNA_PP11_241 | 156108452 | 156108637 |
| LMNA_PP11_465R | 156108638 | 156108657 | |||
| LMNA_PP12_1F | 156108478 | 156108495 | LMNA_PP12_1 | 156108496 | 156108677 |
| LMNA_PP12_225R | 156108678 | 156108763 | |||
| LMNA_PP12_161F | 156108638 | 156108657 | LMNA_PP12_161 | 156108658 | 156108828 |
| LMNA_PP12_371R | 156108829 | 156108848 | |||
| LMNA_PP12_301F | 156108788 | 156108801 | LMNA_PP12_301 | 156108802 | 156108977 |
| LMNA_PP12_519R | 156108978 | 156108997 | |||
| LMNA_PP12_469F | 156108946 | 156108964 | LMNA_PP12_469 | 156108965 | 156109136 |
| LMNA_PP12_682R | 156109137 | 156109159 | |||
3. Results
3.1. Clinical status
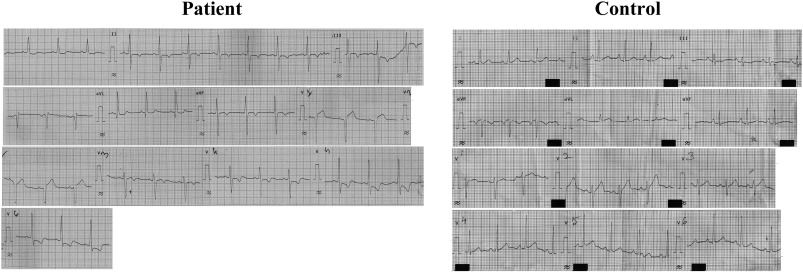
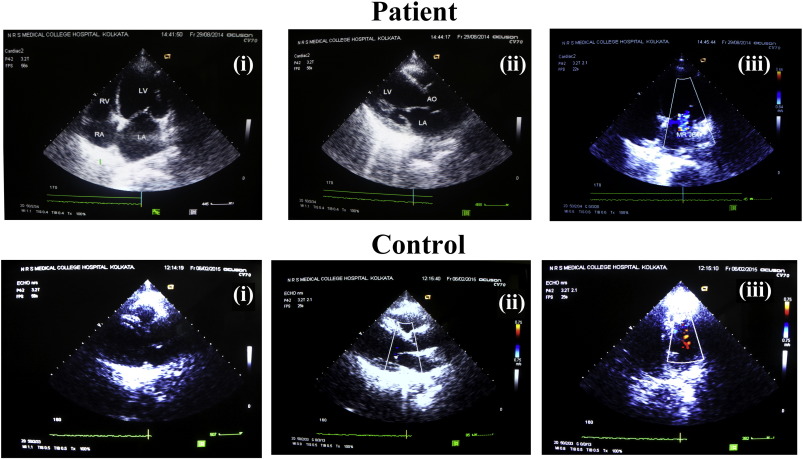
A cohort of 10 subjects coded by S1 through S10, diagnosed with IDCM, from a tertiary care center at Kolkata-N.R.S. Medical College and Hospital were selected randomly irrespective of age and sex. The age group in the cohort varied from 12 to 80 years. The presence of the disease was classified as sporadic in all the patients. Subjects diagnosed with DCM usually showed symptoms of respiratory distress on exertion and also on rest, cough, fatigability and edema. Irregular pulses, narrow pulse pressure, atrial fibrillation, and elevation of jugular venous pressure were routinely observed. Cardiac examinations of the decompensated DCM patients revealed muffled heart sound with gallop rhythm (LVS3 or RVS3). The apex was down and out and there was the presence of systolic murmur at apex which was indicative of mitral regurgitation. Chest X-ray in patients revealed cardiomegaly with or without pulmonary congestion. ECG showed the following abnormalities – LBBB (left bundle branch block), LVH, ST-T and atrial ectopic, ventricular ectopic and also atrial fibrillation as shown in Fig. 1. Echocardiography which is regarded as the gold standard showed dilatation of cardiac chambers, generalized hypokinesia, low ejection fraction and occasional mitral and tricuspid regurgitation as shown in Fig. 2. Detailed clinical statuses of DCM patients are summarized in Table 2.
|
|
|
Fig. 1. A representative ECG of a DCM patient and a control subject. ECG of the patient showed features of LVH with strain pattern. |
|
|
|
Fig. 2. A representative Echocardiogram of a DCM patient and a control subject. ECHO for the patient sample showed the dilation of LV and presence of mitral regurgitation and global hypokinesia. |
| Serial no. | Patient ID | Chest X-ray | ECG | Echocardiography | Coronary angiography | ||
|---|---|---|---|---|---|---|---|
| Dilated cardiac chamber | Global hypokinesia | LV ejection fraction | |||||
| 1 | S1 | Cardiomegaly | Sinus tachycardia | √ | √ | 35% | Normal |
| 2 | S2 | Cardiomegaly, pulmonary plethora | Sinus tachycardia, left ventricular hypertrophy | √ | √ | 22% | Normal |
| 3 | S3 | Cardiomegaly, patchy pneumonia | LBBB | √ | √ | 30% | Normal |
| 4 | S4 | Cardiomegaly | LBBB | √ | √ | 28% | Normal |
| 5 | S5 | Cardiomegaly | LBBB | √ | √ | 35% | Normal |
| 6 | S6 | Cardiomegaly | LBBB, complete heart block | √ | √ | 30% | Normal |
| 7 | S7 | Cardiomegaly | LBBB | √ | √ | 20% | Normal |
| 8 | S8 | Cardiomegaly, patchy pneumonia | LBBB | √ | √ | 28% | Normal |
| 9 | S9 | Cardiomegaly | 2:1 AV block | √ | √ | 32% | Normal |
| 10 | S10 | Cardiomegaly | LBBB | √ | √ | 30% | Normal |
| 11 | Control | No cardiomegaly | Normal | Normal cardiac chamber | Good LV systolic function | 60%–65% | Normal |
3.2. Genetic analysis of LMNA
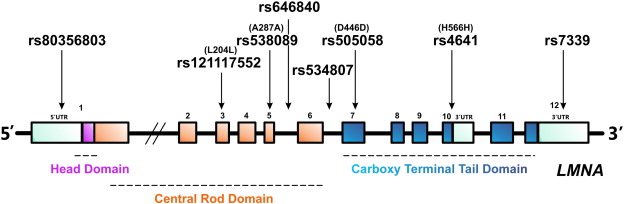
Genetic analysis of the subjects was performed by screening 12 coding exons as well as the immediate intronic regions of LMNA gene, which were amplified by PCR using 29 sets of primers as detailed in Table 1. Using next generation sequencing technique, we identified the association of 8 different LMNA SNPs with 5 out of 10 DCM subjects in our study. All the subjects were found to be heterozygous for the identified SNPs. The identified LMNA SNPs (as shown in Table 3) were (a) rs121117552 c.612G>A L204L located at exon 3, (b) rs538089 c.816T>C A287A located at exon 5, (c) rs505058 c.1338T>C D446D located at exon 7, (d) rs4641 c.1698 C>T H566H located at exon 10 (e) rs646840 c.937-83G>T located in the intron region, (f) rs534807 c.1157+16G>A also residing in the intron region, (g) rs80356803 c.128T>C located in 5′UTR and (h) rs7339 c.76G>C located in the 3′UTR (Fig. 3). Out of the 10 patients surveyed, we observed 4 cases viz. S1, S3, S7 and S9 with multiple LMNA SNPs. Patient S1 scored maximum for 3 SNPs rs505058, rs646840 and rs534807 and remaining 3 patients with 2 SNPs each. However, SNP rs534807 was also identified in another patient named S3. Patient S3 also harbored rs4641 ( Table 3). rs4641 was the most frequently encountered SNP in this cohort, which was identified in three different individuals S3, S4 and S9 (Table 3). SNP rs121117552 was identified in one single patient named S8; rs538089 and rs7339 was found once in patient S7; rs80356803 along with rs4641 were found once in S9 (Table 3). All these SNPs were absent in 12 healthy controls except rs4641 which was identified in one control individual. The loci of the SNPs have been depicted in the schematic diagram of the protein demarcating their respective positions (Fig. 3).
| dbSNP | Amplicon ID | Position of SNP in amplicons | Exon | Type | Zygosity | Genotype | Ref | Variant | Var freq | Ref coverage | Var coverage | Location | Translation impact | Protein variant | Patient ID |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs121117552 | LMNA_PP3_1 | 167 | 3 | SNP | Het | G/A | G | A | 44.44444 | 130 | 104 | Exon | L204L | WT | S8 |
| rs538089 | LMNA_PP5_1 | 144 | 5 | SNP | Het | T/C | T | C | 60.71429 | 55 | 85 | Exon | A287A | WT | S7 |
| rs505058 | LMNA_PP7_81 | 135 | 7 | SNP | Het | T/C | T | C | 55.69493 | 883 | 1110 | Exon | D446D | WT | S1 |
| rs4641 | LMNA_PP10_147 | 82 | 10 | SNP | Het | C/T | C | T | 51.00671 | 292 | 304 | Exon | H566H | WT | S3,S4 & S9 |
| rs646840 | LMNA_PP6_204 | 221 | NA | SNP | Het | G/T | G | T | 81.12642 | 315 | 1354 | Intron | S1 | ||
| rs534807 | LMNA_PP6_204 | 221 | NA | SNP | Het | G/A | G | A | 6.190476 | 197 | 13 | Intron | S1 & S3 | ||
| rs80356803 | LMNA_PP1_1 | 56 | NA | SNP | Het | T/C | T | C | 7.54717 | 49 | 4 | Utr_5 | S9 | ||
| rs7339 | LMNA_PP12_301 | 209 | NA | SNP | Het | G/C | G | C | 50.65327 | 982 | 1008 | Utr_3 | S7 |
Het SNPs = number of called heterozygous SNPs in target regions or loci, ref = genotype in the reference gene, variant = genotype obtained in the patient, var freq = frequency of the variant allele, ref coverage or var coverage = the total reads covering the position, location = position in the gene.
|
|
|
Fig. 3. Schematic representation of 12 exons of LMNA. The SNPs identified in our genetic analysis are displayed at their respective position in the gene with arrows. |
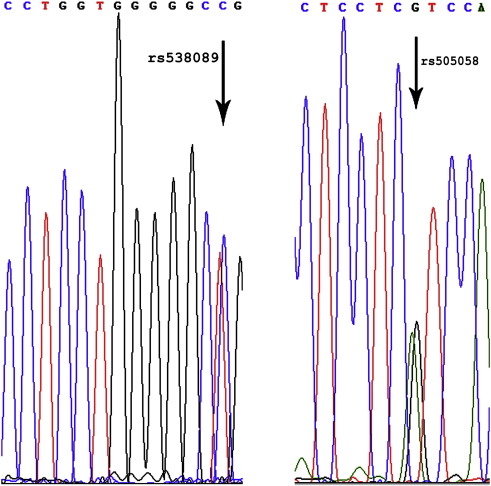
Results obtained from next generation sequencing were further validated by Sanger sequencing. The desired exons of LMNA were amplified by PCR as reported in Perrot et al. [ 9] and were sent for Sanger sequencing. The SNPs residing in the exons of LMNA were all confirmed from the Sanger sequencing, 2 representative chromatograms showing the presence of polymorphism at the desired region are shown in Fig. 4.
|
|
|
Fig. 4. A representative chromatogram of 2 SNPs identified in DCM patients validated by Sanger sequencing; rs538089 validated using forward primer for exon 5; rs505058 validated using reverse primer for exon 7. |
4. Discussion
Laminopathies are principally caused by sporadic mutations in LMNA gene in individuals. Nevertheless, numerous SNPs have been reported from population based studies which are known to cause specific laminopathies like Charcot–Marie–Tooth Disorder, familial partial Lipodystrophy and Emery–Dreifus Muscular dystrophy (EDMD) [17] ; [18]. 165 LMNA mutations have been known to be associated with DCM till date based on studies from Europe, USA and some South East Asian countries but, there are no reports till date establishing any association of SNPs in LMNA with IDCM barring one [25]. However, in a recent report from a Genome Wide Association Study (GWAS) with DCM patients identified two different loci 1) rs10927875 and 2) rs2234962. rs10927875 maps to a region on chromosome 1p36.13. This region encompasses several genes among which HSPB7 was previously suggested to be implicated in DCM whereas rs2234962 was a non-synonymous SNP (c.T757C, p. C151R) which is located within the sequence of BAG3 on chromosome 10q26 [26]. To date, there are very few reports regarding the genetic background of LMNA in Indian population [27] ; [28]. In the present study, we showed for the first time the association of variations in LMNA gene with IDCM in Indian population. We identified 8 LMNA SNPs in subjects with IDCM in a small cohort of Indian sub-population. All these SNPs were shown to be associated with various other disorders in ethnically distinct population but not with DCM.
We identified 4 different SNPs which reside in the exon region of LMNA; the affected exons are 3, 5, 7 and 10 at codon positions 204, 287, 446 and 566 respectively. SNP rs12117552 located in exon 3 of LMNA which was identified in one patient of our study was first reported to be associated with Werners syndrome in an African-American female [29]. rs12117552 in exon 3 which corresponds to silent substitution from G to A at the third base of codon 204 results in same sense variation (rs121117552; L204L). This exon also harbors a mutation hotspot at codon 190, which is critically linked with DCM [9] ; [10]; and thus justifies the development of DCM in patients harboring this variation. Previous reports state the involvement of SNP rs538089 which mapped at exon 5 in LMNA with both Werners syndrome as well as in a subtype of Charcot–Marie–Tooth disease CMT2B1 in North Western African population [29] ; [30]. SNP rs538089 results from a change of nucleotide from T to C at the third base of codon 287; this codon of LMNA is also reported to be associated with a frame shift mutation which causes DCM [25]. Thus, a simple variation but not mutation at codon 287 of LMNA justifies its association with DCM. Next, we identified SNP rs505058 in our study. In rs505058 an alteration at the third base of codon 446 from T to C results in same sense variation to Aspartic acid at exon 7. In other reports from GWAS this SNP was found to be associated with patients afflicted with Late Onset of Alzheimers disease (LOAD) in elderly males [31] ; [32]. It was also found to be involved with Werners syndrome and CMT2B1. The codon 446 was shown to be involved with a mutation D446V in patients suffering from EDMD [33]. EDMD is a condition characterized by weakness of the muscles used for movement (skeletal muscles) and the heart (cardiac) muscle. Consequently, patients suffering with DCM also show weak cardiac muscles, thus a variation at codon position 446 might contribute to the diseased phenotype of DCM.
The most widely studied LMNA SNP in literature is rs4641, which is also the most frequently encountered SNP in our study. A substitution of nucleotide C to T at the third base of codon 566 (exon 10) results in the development of SNP rs4641 [21]; [29]; [31]; [32] ; [34] which still codes for Histidine. Previous studies in ethnically distinct population have shown its association with Type 2 Diabetes (T2D), the metabolic abnormalities, and obesity-related traits in some [20]; [21]; [34]; [35] ; [36]; but not all studies [37] ; [38]. The position associated with rs4641 is very important as it is located adjacent to a splice site at exon 10; alternative splicing at this site leads to the production of either lamin A or C transcripts or protein. A recent report emphasizes the role of T- allele at position 566 in regulating the lamin A to C mRNA ratio in T2D patients [35]. Other reports have shown decreased level of lamin A mRNA in patients suffering from DCM [39]. Thus, it can be hypothesized that rs4641 might contribute to the DCM phenotype by regulating the lamin A to C mRNA ratio although the fact that this is definitely not to be the sole player leading to the pathogenesis of the disease. Interestingly, rs4641 is further associated with Werners syndrome [29]. This makes rs4641 a candidate gene locus as a biomarker for various kind of disorder along with DCM. The other two SNPs rs534807 and rs7339 were reported to be associated with CMT2B1 a commonly known laminopathy. Most of these SNPs were previously reported to be involved with various kinds of disorders except; rs80356803 and rs646840 which were not reported earlier to be involved with any disorders. We showed for the first time the association of these two SNPs with DCM in Indian population and reside in the intron of LMNA. rs538089, rs505058 and rs4641 are the three SNPs identified in Indian population from our study; were also previously known to be involved with DCM in a French population [25]. However we did not observe any other associated symptoms of laminopathies e.g. muscle degeneration, sensory and motor neuron mediated voluntary movement of muscles or unusual accumulation of adipocytes as observed in EDMD, CMT2B1 and FPLD respectively [40]; [41]; [42] ; [43]. Although all patients were admitted based on symptoms which included cough and respiratory distress, these symptoms are common observations for patients of decompensated Dilated Cardiomyopathy.
Currently, there are no special criteria to distinguish LMNA mutation associated Cardiomyopathy and other forms of idiopathic Cardiomyopathy. Genetic heterogeneity is a hallmark for autosomal dominant DCM–Conduction Defects (DCM–CD) where nearly 40 genes have been uncovered to produce disease phenotype [44]; [45]; [46]; [47] ; [48]. Interestingly majority of these genes encode cytoskeletal and sarcomeric proteins. However, mutations in LMNA coding for lamin A/C, DES coding for Desmin and SCN5A encoding cardiac sodium channel protein are still regarded as primary causes for DCM–CD. Therefore, the genes involved in mechanical force transduction and propagation are mutated thereby precipitating the disease. DCM associated with all levels of conduction system defects like sick sinus syndrome, atrioventricular block or bundle branch blocks could be traced back to mutations in LMNA. The conduction disease defects call for pacemaker implantation in patients with LMNA mutations. In young healthy LMNA carriers, arrhythmias including atrial ectopy, atrial fibrillation, non-sustained ventricular tachycardia and ventricular arrhythmias can be the earliest manifestation of the LMNA mutation prior to chamber dilatation [10] ; [23]. Once DCM is clinically confirmed the management follows standard care for heart failure including ACE inhibitors, beta blockers, diuretics and aldosterone antagonists as per the recommendation of New York Heart Association functional class. However, it is not clear whether early administration of these therapeutic agents prior to confirmation of the disease can modify the aggressive nature of LMNA Cardiomyopathy. As LMNA Cardiomyopathy is diagnosed genetically, the sequencing of exons and intron–exon junctions has been reported in various case studies. Due to the advent of deep sequencing technology these sequencing data present a reliable database for cataloging mutations or SNPs in the LMNA gene which might lead to the disease.
The ultimate goal of medical research is to uncover novel diagnostic and therapeutic modalities which will be of clinical utility. Therefore, detailed genetic analysis of the LMNA gene in the affected patients at the early stage might help in better management and hence therapeutic intervention. Such genetic screening based on LMNA gene could be also extended to the nearest kins of the patient to verify any hereditary penetrance which could be then treated as cases of familial DCM. We conclude that polymorphism in LMNA is one of the major genetic risk factor for the pathogenesis of DCM other than mutation in LMNA.
Funding
AB thanks University Grants Council, Government of India for the fellowship. KSG thanks MMDDA and BARD projects of Department of Atomic Energy, Government of India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
Authors declare no conflict of interest.
Acknowledgment
The authors acknowledge Genotypic Technology, India for analyzing the DNA samples of the patients.
References
- [1] G.W. Dec, V. Fuster; Idiopathic dilated cardiomyopathy; N Eng J Med, 331 (23) (1994), pp. 1564–1575
- [2] B.J. Maron, J.A. Towbin, G. Thiene, C. Antzelevitch, D. Corrado, D. Arnett, et al.; Contemporary definitions and classification of the cardiomyopathies an American heart association scientific statement from the council on clinical cardiology, heart failure and transplantation committee; quality of care and outcomes research and functional genomics and translational biology interdisciplinary working groups; and council on epidemiology and prevention; Circulation, 113 (14) (2006), pp. 1807–1816
- [3] J.A. Towbin, N.E. Bowles; Dilated cardiomyopathy: a tale of cytoskeletal proteins and beyond; J Cardiovasc Electrophysiol, 17 (8) (2006), pp. 919–926
- [4] M. Taylor, E. Carniel, L. Mestroni; Cardiomyopathy, familial dilated; Orphanet J Rare Dis, 1 (1) (2006), p. 27
- [5] R.E. Hershberger, J.D. Siegfried; Update 2011: clinical and genetic issues in familial dilated cardiomyopathy; J Am Coll Cardiol, 57 (16) (2011), pp. 1641–1649
- [6] M.R. Taylor, P.R. Fain, G. Sinagra, M.L. Robinson, A.D. Robertson, E. Carniel, et al.; Natural history of dilated cardiomyopathy due to lamin A/C gene mutations; J Am Coll Cardiol, 41 (5) (2003), pp. 771–780
- [7] R.E. Hershberger, D. Nauman, T.L. Walker, D. Dutton, D. Burgess; Care processes and clinical outcomes of continuous outpatient support with inotropes (COSI) in patients with refractory endstage heart failure; J Card Fail, 9 (3) (2003), pp. 180–187
- [8] P.M. Jakobs, E.L. Hanson, K.A. Crispell, W. Toy, H. Keegan, K. Schilling, et al.; Novel lamin A/C mutations in two families with dilated cardiomyopathy and conduction system disease; J Card Fail, 7 (3) (2001), pp. 249–256
- [9] A. Perrot, S. Hussein, V. Ruppert, H.H. Schmidt, M.S. Wehnert, N.T. Duong, et al.; Identification of mutational hot spots in LMNA encoding lamin A/C in patients with familial dilated cardiomyopathy; Basic Res Cardiol, 104 (1) (2009), pp. 90–99
- [10] E. Arbustini, A. Pilotto, A. Repetto, M. Grasso, A. Negri, M. Diegoli, et al.; Autosomal dominant dilated cardiomyopathy with atrioventricular block: a lamin A/C defect-related disease; J Am Coll Cardiol, 39 (6) (2002), pp. 981–990
- [11] M. Pasotti, C. Klersy, A. Pilotto, N. Marziliano, C. Rapezzi, A. Serio, et al.; Long-term outcome and risk stratification in dilated cardiolaminopathies; J Am Coll Cardiol, 52 (15) (2008), pp. 1250–1260
- [12] P. Sebillon, C. Bouchier, L. Bidot, G. Bonne, K. Ahamed, P. Charron, et al.; Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations; J Med Genet, 40 (8) (2003), pp. 560–567
- [13] Spaendonck-Zwarts KY, I.A. Rijsingen, M.P. Berg, R.H. Lekanne Deprez, J.G. Post, A.M. Mil, et al.; Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years' experience; Eur J Heart Fail, 15 (6) (2013), pp. 628–636
- [14] J.A. Towbin, R.J. Solaro; Genetics of dilated cardiomyopathy: more genes that kill; J Am Coll Cardiol, 44 (10) (2004), pp. 2041–2043
- [15] D. Fatkin, C. MacRae, T. Sasaki, M.R. Wolff, M. Porcu, M. Frenneaux, et al.; Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease; New Eng J Med, 341 (23) (1999), pp. 1715–1724
- [16] F. Lin, H.J. Worman; Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C; J Biol Chem, 268 (22) (1993), pp. 16321–16326
- [17] C.J. Hutchison; Lamins: building blocks or regulators of gene expression?; Nat Rev Mol Cell Biol, 3 (11) (2002), pp. 848–858
- [18] S. Benedetti, L. Merlini; Laminopathies: from the heart of the cell to the clinics; Curr Opin Neurol, 17 (5) (2004), pp. 553–560
- [19] J. Scharner, V. Gnocchi, J. Ellis, P. Zammit; Genotype–phenotype correlations in laminopathies: how does fate translate?; Biochem Soc Trans, 38 (1) (2010), p. 257
- [20] R.A. Hegele, H. Cao, S.B. Harris, B. Zinman, A.J. Hanley, C.M. Anderson; Genetic variation in LMNA modulates plasma leptin and indices of obesity in aboriginal Canadians; Physiol Genomics, 3 (1) (2000), pp. 39–44
- [21] R.A. Hegele, M.W. Huff, T.K. Young; Common genomic variation in LMNA modulates indexes of obesity in Inuit 1; J Clin Endocrinol Metabol, 86 (6) (2001), pp. 2747–2751
- [22] C. Weyer, J.K. Wolford, R.L. Hanson, J.E. Foley, P.A. Tataranni, C. Bogardus, et al.; Subcutaneous abdominal adipocyte size, a predictor of type 2 diabetes, is linked to chromosome 1q21–q23 and is associated with a common polymorphism in LMNA in pima Indians; Mol Genet Metab, 72 (3) (2001), pp. 231–238
- [23] J.H. van Berlo, W.G. de Voogt, A.J. van der Kooi, J.P. van Tintelen, G. Bonne, R.B. Yaou, et al.; Meta-analysis of clinical characteristics of 299 carriers of LMNA gene mutations: do lamin A/C mutations portend a high risk of sudden death?; J Mol Med, 83 (1) (2005), pp. 79–83
- [24] P. Richardson, R. McKenna, M. Bristow, B. Maisch, B. Mautner, J. O'connell, et al.; Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the definition and classification of car diomyopathies; Circulation, 93 (5) (1996), pp. 841–842
- [25] G. Millat, V. Chanavat, S. Julia, H. Crehalet, P. Bouvagnet, R. Rousson; Validation of high-resolution DNA melting analysis for mutation scanning of the LMNA gene; Clin Biochem, 42 (9) (2009), pp. 892–898
- [26] E. Villard, C. Perret, F. Gary, C. Proust, G. Dilanian, C. Hengstenberg, et al.; A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy; Eur Heart J, 32 (9) (2011), pp. 1065–1076
- [27] M. Sharma, A. Misra, N. Vikram, B. Suryaprakash, S. Chhabra, N. Garg, et al.; Genotype of the LMNA 1908C > T variant is associated with generalized obesity in Asian Indians in North India; Clin Endocrinol, 75 (5) (2011), pp. 642–649
- [28] B. Ushasree, V. Shivani, A. Venkateshwari, R. Jain, C. Narsimhan, P. Nallari; Epidemiology and genetics of dilated cardiomyopathy in the Indian context; Indian J Med Sci, 63 (7) (2009), p. 288
- [29] K.N. Jacob, F. Baptista, H.G. dos Santos, J. Oshima, A.K. Agarwal, A. Garg; Phenotypic heterogeneity in body fat distribution in patients with atypical Werners syndrome due to heterozygous Arg133Leu lamin A/C mutation; J Clin Endocrinol Metabol, 90 (12) (2005), pp. 6699–6706
- [30] T. Hamadouche, Y. Poitelon, E. Genin, M. Chaouch, M. Tazir, N. Kassouri, et al.; Founder effect and estimation of the age of the c. 892C > T (p. Arg298Cys) mutation in LMNA associated to Charcot–Marie–Tooth subtype CMT2B1 in families from North Western Africa; Ann Hum Genet, 72 (5) (2008), pp. 590–597
- [31] C. Cluett, C. Brayne, R. Clarke, G. Evans, F. Matthews, D.C. Rubinsztein, et al.; Polymorphisms in LMNA and near a SERPINA gene cluster are associated with cognitive function in older people; Neurobiol Aging, 31 (9) (2010), pp. 1563–1568
- [32] H.-L. Yeh, S.-J. Hou, F.-C. Yen, C.-J. Hong, Y.-J. Liou, A.C. Yang, et al.; Polymorphisms in LMNA and near a SERPINA13 gene are not associated with cognitive performance in Chinese elderly males without dementia; Neurosci Lett, 504 (1) (2011), pp. 32–34
- [33] L. Spinarova, J. Toman, P. Hude, S. Vohanka, M. Vytopil, Z. Lukas, et al.; [Disorders of laminins in diseases of myocardial and skeletal muscles]; Vnitr Lek, 49 (8) (2003), pp. 637–641
- [34] N.I. Steinle, R. Kazlauskaite, I.G. Imumorin, W.-C. Hsueh, T.I. Pollin, J.R. O'Connell, et al.; Variation in the Lamin A/C gene associations with metabolic syndrome; Arterioscler Thromb Vasc Biol, 24 (9) (2004), pp. 1708–1713
- [35] L. Wegner, S. Anthonsen, J. Bork-Jensen, L. Dalgaard, T. Hansen, O. Pedersen, et al.; LMNA rs4641 and the muscle lamin A and C isoforms in twins—Metabolic implications and transcriptional regulation; J Clin Endocrinol Metabol, 95 (8) (2010), pp. 3884–3892
- [36] L. Wegner, G. Andersen, T. Sparsø, N. Grarup, C. Glümer, K. Borch-Johnsen, et al.; Common variation in LMNA increases susceptibility to type 2 diabetes and associates with elevated fasting glycemia and estimates of body fat and height in the general population studies of 7,495 Danish Whites; Diabetes, 56 (3) (2007), pp. 694–698
- [37] J.L. Mesa, R.J. Loos, P.W. Franks, K.K. Ong, et al.; Lamin A/C polymorphisms, type 2 diabetes, and the metabolic syndrome case–control and quantitative trait studies; Diabetes, 56 (3) (2007), pp. 884–889
- [38] K. Duesing, G. Charpentier, M. Marre, J. Tichet, S. Hercberg, P. Froguel, et al.; Evaluating the association of common LMNA variants with type 2 diabetes and quantitative metabolic phenotypes in French Europids; Diabetologia, 51 (1) (2008), pp. 76–81
- [39] N. Narula, V. Favalli, P. Tarantino, M. Grasso, A. Pilotto, R. Bellazzi, et al.; Quantitative expression of the mutated lamin A/C gene in patients with cardiolaminopathy; J Am Coll Cardiol, 60 (19) (2012), pp. 1916–1920
- [40] G. Bonne, M.R. Di Barletta, S. Varnous, H.-M. Bécane, E.-H. Hammouda, L. Merlini, et al.; Mutations in the gene encoding lamin A/C cause autosomal dominant Emery–Dreifuss muscular dystrophy; Nat Genet, 21 (3) (1999), pp. 285–288
- [41] A. De Sandre-Giovannoli, M. Chaouch, S. Kozlov, J.-M. Vallat, M. Tazir, N. Kassouri, et al.; Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot–Marie–Tooth disorder type 2) and mouse; Am J Hum Genet, 70 (3) (2002), pp. 726–736
- [42] H. Cao, R.A. Hegele; Nuclear lamin A/C R482Q mutation in Canadian kindreds with Dunnigan-type familial partial lipodystrophy; Hum Mol Genet, 9 (1) (2000), pp. 109–112
- [43] S. Shackleton, D.J. Lloyd, S.N. Jackson, R. Evans, M.F. Niermeijer, B.M. Singh, et al.; LMNA, encoding lamin A/C, is mutated in partial lipodystrophy; Nat Genet, 24 (2) (2000), pp. 153–156
- [44] T.J. Cahill, H. Ashrafian, H. Watkins; Genetic cardiomyopathies causing heart failure; Circ Res, 113 (6) (2013), pp. 660–675
- [45] P. Teekakirikul, M.A. Kelly, H.L. Rehm, N.K. Lakdawala, B.H. Funke; Inherited cardiomyopathies: molecular genetics and clinical genetic testing in the postgenomic era; J Mol Diagn, 15 (2) (2013), pp. 158–170
- [46] M.J. Ackerman, S.G. Priori, S. Willems, C. Berul, R. Brugada, H. Calkins, et al.; HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA); Europace, 13 (8) (2011), pp. 1077–1109
- [47] A. Morales, R.E. Hershberger; Genetic evaluation of dilated cardiomyopathy; Curr Cardiol Rep, 15 (7) (2013), pp. 1–8
- [48] N. Campbell, G. Sinagra, K.L. Jones, D. Slavov, K. Gowan, M. Merlo, et al.; Whole exome sequencing identifies a troponin T mutation hot spot in familial dilated cardiomyopathy; PLoS One, 8 (10) (2013), p. e78104
Document information
Published on 19/05/17
Submitted on 19/05/17
Licence: Other
Share this document
Keywords
claim authorship
Are you one of the authors of this document?