(Created page with " ==Highlights== * HOTAIR and MALAT1 were upregulated in PBLCs of PAHs-exposed workers. * HOTAIR and MALAT1 expression was positively associated with the degree of DNA damage...") |
m (Scipediacontent moved page Draft Content 352853087 to Gao et al 2016a) |
(No difference)
| |
Revision as of 11:27, 2 May 2017
Highlights
- HOTAIR and MALAT1 were upregulated in PBLCs of PAHs-exposed workers.
- HOTAIR and MALAT1 expression was positively associated with the degree of DNA damage induced by PAHs.
- H3K27me3 modification was positively correlated with the degree of genetic damage and the increase of HOTAIR expression.
Abstract
To explore whether the alteration of lncRNA expression is correlated with polycyclic aromatic hydrocarbons (PAHs) exposure and DNA damage, we examined PAHs external and internal exposure, DNA damage and lncRNAs (HOTAIR, MALAT1, TUG1 and GAS5) expression in peripheral blood lymphocytes (PBLCs) of 150 male coke oven workers and 60 non-PAHs exposure workers. We found the expression of HOTAIR, MALAT1, and TUG1 were enhanced in PBLCs of coke oven workers and positively correlated with the levels of external PAHs exposure (adjusted Ptrend < 0.001 for HOTAIR and MALAT1, adjusted Ptrend = 0.006 for TUG1). However, only HOTAIR and MALAT1 were significantly associated with the level of internal PAHs exposure (urinary 1-hydroxypyrene) with adjusted β = 0.298, P = 0.024 for HOTAIR and β = 0.090, P = 0.034 for MALAT1. In addition, the degree of DNA damage was positively associated with MALAT1 and HOTAIR expression in PBLCs of all subjects (adjusted β = 0.024, P = 0.002 for HOTAIR and β = 0.007, P = 0.003 for MALAT1). Moreover, we revealed that the global histone 3 lysine 27 trimethylation (H3K27me3) modification was positively associated with the degree of genetic damage (β = 0.061, P < 0.001) and the increase of HOTAIR expression (β = 0.385, P = 0.018). Taken together, our findings suggest that altered HOTAIR and MALAT1 expression might be involved in response to PAHs-induced DNA damage.
Abbreviations
PAHs , polycyclic aromatic hydrocarbons ; PBLCs , peripheral blood lymphocytes ; 1-OHP , 1-hydroxypyrene ; lncRNAs , long non-coding RNAs ; HOTAIR , HOX transcript antisense RNA ; MALAT1 , metastasis-associated lung adenocarcinoma transcript 1 ; TUG1 , taurine up-regulated 1 ; GAS5 , growth arrest-specific 5 ; H2K27me3 , histone 3 lysine 27 trimethylation
Chemical compound
1-hydroxypyrene (PubChem CID: 21387)
Keywords
Polycyclic aromatic hydrocarbons ; Long non-coding RNA ; Peripheral blood lymphocytes ; DNA damage response ; HOTAIR ; MALAT
1. Introduction
PAHs are ubiquitous occupational and environmental contaminants, which have been classified as human genotoxicants and carcinogens [15] and [51] . Epidemiological studies have demonstrated that long-term exposure to PAHs links to high incidence of lung cancer in coke oven workers [5] . As a complex disease, the molecular etiology of cancer includes both genetic modifications and epigenetic aberrations. In genetics, DNA damage, oncogene activation and inactivation of tumor suppressor gene are believed to play important roles in PAHs-induced carcinogenesis [16] . In epigenetics, aberrant DNA methylation, histone modification and miRNA expression patterns have currently emerged as important mechanisms that contribute to PAHs-induced genotoxicity and carcinogenicity [6] , [7] , [24] , [32] and [46] . Long non-coding RNAs (lncRNA) is one of the essential epigenetic regulators. The specificity of lncRNA expression is now recognized as important epigenetic marks that confers lncRNAs with great potential as biomarkers for health risk assessment [29] . Accumulating evidence demonstrate that lncRNAs play critical roles not only in physiological processes of normal cells, but also in the development of many kinds of human diseases [10] , [29] , [33] , [36] , [43] , [45] and [48] . Recent studies found that alteration of lncRNA expression was involved in induction of several chemicals-induced genotoxicity and cell malignant transformation, indicating the aberrant lncRNA changes might be promising biomarkers for risk prediction of environmental exposure [3] , [9] , [13] , [26] , [31] and [47] . However, how ambient PAHs exposure affects lncRNA expression and whether it is involved in development of adverse health effects have not been defined.
In this study, four candidate lncRNAs, HOTAIR, MALAT1, TUG1 and GAS5 that have been reported to be associated with DNA damage and cancer development [8] , [12] , [14] , [17] , [21] , [22] , [28] , [30] , [34] , [37] , [38] , [39] , [40] , [49] and [50] were selected to address the biological significance of lncRNAs. Our findings reveal that the altered HOTAIR and MALAT1 expression could be the sensitive biomarkers that indicate the PAHs exposure and PAHs-induced DNA damage.
2. Materials and methods
2.1. Study population and sample collection
In this study, 150 PAHs-exposure workers in coking plant and 60 non-PAHs exposure workers in hot-rolling mill from Ben Xi Iron and Steel Group Cooperation in Liaoning Province, China were recruited. Those workers who had suffered from acute infectious diseases, chronic diseases, long-term drug use, or exposed to mutagenic agents (such as X-ray radiation) within 2 months were excluded. Basic information of each subject was collected by a structured epidemiologic questionnaire including demographic information, educational level, smoking history, alcohol consumption, occupational exposure history, personal medical history, and grilled food intake. Additionally, 15 mL urine was collected for urinary 1-hydroxypyrene (1-OHP) detection and 5 mL of venous blood were drawn in an EDTA-Na2 containing tube for comets assay and PBLCs isolation. PBLCs were isolated by using a standardized Ficoll-Hypaque gradient procedure in less than 3 h after the blood samples were obtained. All samples were kept at −80 °C before analysis. The protocol was approved by Research Ethic Committee of School of Public Health, Sun Yat-sen University, and informed consent was obtained from each participant.
2.2. Urine 1-OHP detection
5 mL of 4-day shift-end urine were collected and the measurement of 1-hydroxypyrene (1-OHP) were carried out according to the method described previously [19] . The level of urinary 1-OHP was detected by high-pressure liquid chromatography (HPLC) equipped with a fluorescence spectrophotometer and normalized by urinary creatinine (Cr) and presented as microgram per gram creatinine. The detection limit was 0.14 μg/L urine (signal/noise = 3). Measurements below the limit of detection (LOD) were replaced with LOD/ .
2.3. Alkaline comets assay
The comet assay was performed using method described by Li et al. [24] and Singh et al. [35] . Comet assay was performed using the fresh blood sample according to the protocol. In order to minimize the variation, we placed the same numbers of samples collected from different groups (control, bottom, side and top groups) in one slide (CometSlide HT, Trevigen, USA). Analyses of the images were performed using Comet Assay Software Project-1.2.2 (University of Wroclaw, Poland). Olive tail moment (OTM) was selected as the parameter to indicate the degree of DNA damage.
2.4. RNA isolation and qRT-PCR
Total RNA was isolated using TRIzol Reagent (Invitrogen, Carlsbad, USA) and quantitative reverse transcription was carried out using TOYOBO RT-qPCR Kit (TOYOBO, Tokyo, Japan). The mRNA expression was detected using the SYBR Green Real time PCR Master Mix-Plus (TOYOBO, Tokyo, Japan) with the standard program in a Viia 7 Real-time PCR system (Applied Biosystems, Branford, USA). The amplification of GAPDH was used as an internal control to normalize lncRNA expression. The sequences of qRT-PCR primers were listed in Supplementary Table 1. RNA sample isolated from 16HBE cells was used for internal quality control among different experiments. In particular, specific gene expression in RNA samples of 16HBE cells from each run of qPCR were adjusted by both ΔCtbatchbalance (Ctbatchtargetgene − CtbatchGAPDH ) and CtGAPDH . The expression of lncRNA was calculated using an equation of ΔCt = Cttarge gene − CtGAPDH − ΔCtbatchbalance . All results were expressed as mean ± SD from three independent experiments.
2.5. Histone extraction and detection of H3K27me3 modification
The histone extraction was performed following a protocol described previously [2] and [4] . During the process of isolation, the protease inhibitor mixture (Roche Applied Sciences, Indianapolis, USA) and 1 mM phenylmethylsulfonyl fluoride were used for protein stabilization. The histone precipitation was dried by the air and dissolved in sterile deionized water before detection. Subsequently, sandwich enzyme-linked immunosorbent (ELISA) assay methods were used for determining the levels of histone modification. ELISA was conducted in a 96-well plate coated with 1:20000 histone H3 antibody (Abcam, Cambridge, USA) overnight at 4 °C. Samples and H3K27me3 recombinant proteins (Active Motif, Carlsbad, USA) were added to the plates and incubated for 1.5 h. Then the primary antibodies including histone H3 at 1:10000 (Sigma–Aldrich, USA) or H3K27me3 at 1:3000 (Abcam, Cambridge, USA) were added to each well separately and incubated for 2 h, following by an addition of secondary antibody at 1:1000 dilution (Santa Cruz Biotechnology, Santa Cruz, USA). After 2 h incubation, 3,3′,5,5″-tetramethylbenzidine (Beyotime, Suzhou, China) solution was added and incubated for additional 30 min in the dark. The reaction was stopped by addition of 30 μL of 2 M H2 SO4 and the optical density was read at 450 nm using a microplate spectrophotometer (BIO-TEC, Vermont, USA). Data was presented as mean ± SD from triplicate wells of two independent experiments.
2.6. Statistical analyses
In the current analysis we examined the data for normality (Shapiro–Wilks W -test) and homogeneity (Bartletts test for unequal variances). Continuous variables are provided as the mean ± SD, while categorical variables are displayed as the percentage within each subgroup. Potential associations between categorical variables were assessed by contingency tables and the χ2 -test. Comparisons between continuous variables between groups were performed by analysis of t -test. The Pearson correlation coefficient was used to quantify the correlation between two factors. Multiple linear regression analysis was used to correct the confounders by a stepwise regression method. The adjusted β value and P value was obtained from AIC (Akaike Information Criterion) lowest models. The non-normal distribution values were logarithmic transformed before regression analysis. All P values were two sided, with only P < 0.05 considered statistically significant. All statistical analyses were using the base packages in R version (R -projecting.org. (C) 1998–2012).
3. Results
3.1. Population characteristics and PAHs exposure levels
In this study, we recruited 150 PAHs-exposure workers in coking plant and 60 non-PAHs exposure workers in hot-rolling mill from an Iron and Steel Group Cooperation (Table 1 ). There were no significant differences between PAHs-exposed group and control group in terms of smoking status, drinking status, education, and BMI. Factors such as age, ethic and grill food consumption differed significantly between two group subjects. Remarkably, the internal exposure biomarker urinary 1-OHP in exposure groups was 6-fold higher than that in control group (P < 0.001).
| Controls (n = 60) | PAHs-exposed workers (n = 150) | P -value | |
|---|---|---|---|
| General characteristic | |||
| Age (years) | 46.38±8.97 | 42.20±7.38 | 0.002a |
| Coke history(years) | – | 16.29±7.75 | – |
| Smokers [n (%)] | 39 (65.00) | 97 (64.67) | 0.963b |
| Smoking age (years) | 15.39±14.42 | 12.73±11.42 | 0.208a |
| Drinkers [n (%)] | 38 (63.00) | 88 (58.67) | 0.533b |
| Education (years) | 0.662b | ||
| ≤9 | 30 (50.00) | 70 (46.67) | |
| >9 | 30 (50.00) | 80 (53.33) | |
| BMI | 24.93±3.88 | 24.68±2.95 | 0.657a |
| Ethic (Han/others) | 58 (96.67) | 120 (80.00) | 0.002b |
| Grill food consumption [n (%)] | 0 (0.00) | 50 (66.67) | <0.001b |
| Internal exposure biomarker | |||
| Urinary 1-OHP (μg/g creatinine) | 7.09 (4.81,10.00) | 43.70 (12.99,136.18) | <0.001c |
| Lnc RNA expression (−ΔCT) d | |||
| HOTAIR | −13.21 (−11.80,−14.37) | −11.79 (−10.88,−12.81) | <0.001c |
| TUG1 | −5.02 (−4.61,−5.32) | −4.20 (−3.48,−5.00) | <0.001c |
| MALAT1 | −0.82 (−0.37,−1.27) | 0.15 (0.74,−0.46) | <0.001c |
| GAS5 | 0.72 (1.23,0.29) | 1.02 (1.43,0.50) | 0.495c |
| DNA damage | |||
| Olive tail moment | 3.27 (0.00,5.54) | 40.76 (4.86,53.61) | <0.001c |
| Histone modification | |||
| H3K27me3 (%) | 9.63±3.82 | 12.50±7.15 | 0.030c |
Statistical significant indexes are in Bold.
a. Two-sides Student t test.
b. Two-sides chi-square test.
c. Multivariate covariance analysis with adjustment for BMI, age, smoke status, drinking status, grill food consumption, education, and ethics.
d. The ΔCT = CTtarget − CTGAPDH − ΔCTbatchbalance .
3.2. The alteration of lncRNA expression in response to PAHs exposure
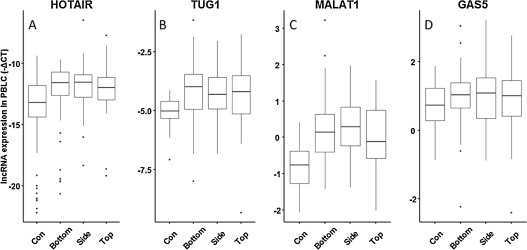
The expression patterns of four selected lncRNAs (HOTAIR, MALAT1, TUG1 and GAS5) were examined in PBLCs of all subjects. As shown in Table 1 , the expression of HOTAIR, MALAT1 and TUG1was 2.68-, 1.96-, and 1.77-fold, respectively higher in PAHs-exposed group compared to control group (P < 0.001). In addition, when we divided PAHs-exposed group into three subgroups as bottom, side, and top oven, respectively based on the working distance to the coke oven where the level of PAHs exposure went up from bottom to top as previously described [24] . As shown in Fig. 1 , we found that the expression of HOTAIR (adjusted Ptrend < 0.001), TUG1 (adjusted Ptrend = 0.006) and MALAT1 (adjusted Ptrend < 0.001) in PBLCs were upregulated with the increase of PAHs exposure analyzed by trend analysis after adjustment for confounder factors such as BMI, age, smoke status, drinking status, grill food consumption, education, and ethics (Supplemental Table 2).
|
|
|
Fig. 1. Association between PAHs exposure and lncRNA expression in PBLCs of the subjects. The PAHs-exposed workers were divided into three subgroups as bottom, side, and top oven, where the level of PAHs exposure went up from bottom to top. LncRNA expression was tested by qRT-PCR method. The horizontal line in the box represents the median of each subgroup. (A) HOTAIR, Ptrend < 0.001; (B) TUG1, Ptrend = 0.002; (C) MALAT1, Ptrend < 0.001; (D) GAS5, Ptrend = 0.809. |
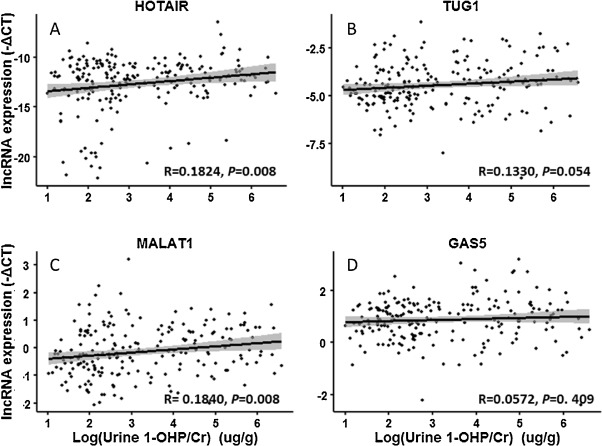
Next, we clarified the correlation between the internal biomarker (1-OHP) and lncRNAs expression in PBLCs of the subjects through stepwise multiple regression analysis after adjustment for factors described above. As shown in Fig. 2 , the expression of HOTAIR and MALAT1 (both P = 0.008) was positively associated with urinary 1-OHP, while TUG1 (P = 0.054) and GAS5 (P = 0.409) were not statistically correlated with urinary 1-OHP. Moreover, we found that the concentration of urinary 1-OHPwas positive correlated with the levels of HOTAIR (β = 0.298, P = 0.024) and MALAT1 (β = 0.090, P = 0.034; Table 2 and Supplemental Table 3). These findings reveal that altered HOTAIR and MALAT1 expression in PBLCs of coke oven workers could be an indicator of PAHs exposure.
|
|
|
Fig. 2. The correlation between urinary 1-OHP and lncRNA expression in PBLCs of all subjects. LncRNA expression levels (Y -axis) were presented as −ΔCt = −(Cttargetgene − CtGAPDH − ΔCtbatchbalance ). Urinary 1-OHP (X -axis) was presented as log-transformed value. (A) HOTAIR, y = 0.3431x − 13.7423, R = 0.1824, P = 0.008; (B) TUG1, y = 0.1086x − 4.7992, R = 0.1330, P = 0.054; (C) MALAT1, y = 0.1134x − 0.5052, R = 0.1840, P = 0.008; (D) GAS5, y = 0.0001x − 0.8388,R = 0.0572,P = 0. 409. |
| Urinary 1-OHPa | ||||
|---|---|---|---|---|
| Unadjusted | Adjustedb | |||
| β | P -value | β | P -value | |
| LncRNA expression (−ΔCT) | ||||
| HOTAIR | 0.343 | 0.008 | 0.298 | 0.024 |
| TUG1 | 0.109 | 0.054 | 0.110 | 0.060 |
| MALAT1 | 0.113 | 0.008 | 0.090 | 0.034 |
| GAS5 | <0.001 | 0.409 | 0.065 | 0.265 |
| DNA damage | ||||
| Olive tail moment | 4.991 | <0.001 | 4.657 | <0.001 |
Statistical significant indexes are in Bold.
a. Urinary 1-OHP was normalized by urinary creatinine (Cr) and the values were logarithmic transformation before regression analysis.
b. Multiple linear regression analysis was performed for P value. Analysis adjusted by BMI, age, smoke status, drinking status, grill food consumption, education, and ethics. The details are presented in Supplementary Table 3.
Next, we analyze the DNA damage of PBLCs in all subjects using Comet assay and selected OTM as the parameter indicating the degree of DNA damage. Similar to our previous findings, the degree of DNA damage in PAHs-exposed group was increased by12.5-fold compared to the control group (Table 1 ). In addition, results from linear and multiple regression analysis (Table 2 ) showed that the degree of DNA damage was significantly associated with the levels of 1-OHP (adjusted β = 4.657, P < 0.001). In addition, we found that the levels of HOTAIR and MALAT1 were positively correlated with the degree of DNA damage (HOTAIR: β = 2.156, P < 0.001; MALAT1: β = 6.621, P < 0.001). In contrast, no association was observed between the expression of TUG1, GAS5 and the extent of DNA damage (Table 3 ). These results indicate that HOTAIR and MALAT1 might be involved in regulation of DNA damage in response to PAHs exposure.
| Lnc RNA expression (−ΔCT) | Olive tail moment | |||
|---|---|---|---|---|
| Unadjusted | Adjusteda | |||
| β | P -value | β | P -value | |
| HOTAIR | 0.028 | <0.001 | 0.024 | 0.002 |
| TUG1 | 0.004 | 0.216 | 0.002 | 0.641 |
| MALAT1 | 0.009 | <0.001 | 0.007 | 0.003 |
| GAS5 | 0 | 0.919 | 0 | 0.904 |
Statistical significant indexes are in Bold.
a. Multiple linear regression analysis was performed. Analysis adjusted by BMI, age, smoke status, drinking status, grill food consumption, education, and ethics.
3.4. The association of HOTAIR expression and H3K27me3 modification in PBLCs of the subjects
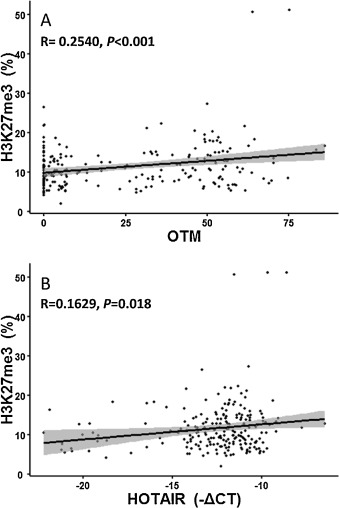
H3K27me3 had been considered as a biomarker in response to DNA damage [18] and [25] and could be upregulated by HOTAIR and polycomb group (PcG) protein complexes in in vitro studies [11] and [41] . Therefore, we analyzed the correlation of H3K27me3 modification with DNA damage and the HOTAIR expression in PBLCs of all subjects. As a result, H3K27me3 modification were elevated by 29.80% in PBLCs of coke oven workers compared to that of control workers (P < 0.030, Table 1 ). Consistently, we found that the extent of DNA damage (β = 0.061, P < 0.001) and the expression of HOTAIR (β = 0.385, P = 0.018) were both positively associated with H3K27me3 modifications analyzed by a linear regression model (Fig. 3 ). Taken together, these findings indicate that lncRNA may regulate cellular response to DNA damage through interplay with histone modification.
|
|
|
Fig. 3. Enhanced H3K27me3 modification was correlated to the degree of DNA damage and upregulation of HOTAIR expression. The modification of H3K27me3 was examined by ELISA. The degree of DNA damage was indicated by Olive tail moment (OTM) from Comet assay. LncRNA expression was tested by qRT-PCR. (A) Association of H3K27me3 modification with OTM in PBLCs of all subjects. y = 0.0606x + 9.9009, R = 0.2540, P < 0.001. (B) Association of H3K27me3 modification with HOTAIR expression in PBLCs of all subjects. y = 0.0182x + 16.5407, R = 0.1629, P = 0.018. |
4. Discussion
Studies of lncRNAs in human populations could identify biomarkers of exposure and/or early effect and elucidate mechanisms of action underlying environmental exposure associated diseases. Occupational exposure to PAHs has been shown in in vitro studies to alter the expression of non-coding RNA. In this study, we attempt to address how lncRNA responses to environmental stress in the context of population exposure. Here, we reveal that two lncRNAs, HOTAIR and MALAT1 whose aberrant expression was positively associated with the PAHs exposure and DNA damage.
LncRNAs have been reported to be involved in multiple biological processes and human diseases through transcriptional, posttranscriptional and epigenetic regulation [20] . Recent studies showed that a number of lncRNAs mediated the malignant transformation of human bronchial epithelial cells induced by benzo(a) pyrene (Bap) and its ultimate carcinogen, anti-benzopyrene-trans-7,8-dihydrodiol-9,10-epoxide (anti-BPDE) through regulating cell apoptosis and proliferation [9] , [13] and [47] . In this study, we found that the expression of HOTAIR, MALAT1 and TUG1 were significantly up-regulated in PAHs-exposed workers. Of these three lncRNAs, HOTAIR and MALAT1 were positively associated with exposure marker, implying a role of lncRNA in mediating environmental stress-induced adverse health effects. Similar results were found in an in vitro study showing that HOTAIR and MALAT1 were upregulated in HBE cells treated by cigarette smoke extract (CSE) for 20 weeks. In addition, they found that HOTAIR might play an important role in epithelial-mesenchymal transition and the formation of stem cell-like properties [26] . Taken together, these observations suggested that HOTAIR and MALAT1 might be applicable in the surveillance of epigenetic damage for environmental exposure.
Although previous studies revealed that lncRNAs might be involved in chemical-induced genotoxicity [27] and [31] , the association between lncRNA changes and DNA damage had not been clarified, particularly in population-based studies. In this study, we demonstrate that altered MALAT1 and HOTAIR expression in PBLCs of PAHs exposed workers are correlated with the degree of DNA damage, indicating their roles in predicting PAHs-induced genotoxicity. MALAT1is a highly abundant and ubiquitously expressed long noncoding RNA (lncRNA), which may predict prognosis in several types of cancers [42] . Previous study revealed that MALAT1 is a key regulator that promotes the cell cycle by inhibiting p53 activation and regulating the expression of B-Myb [39] . Similarly, another research group also found that MALAT1 was upregulated in both TK6 and WTK1 cells after the ionizing radiation-induced DNA damage [1] . Taken together, these findings suggest that MALAT1 may be involved in mediating the response to PAHs-induced DNA damage.
Previous studies uncover an oncogenic role of HOTAIR in promoting proliferation, migration and invasion of cancer cells [8] , [37] and [44] . Moreover, HOTAIR-mediated cancer development is tightly associated with a specific histone modification, H3 lysine27 trimethylation (H3K27), resulting in a chromatin state that prevents the transcription of several tumor suppressor genes [11] . Recent study found that the global H3K27me3 level was increased in human cells after chemicals treatment or UV radiation [18] and [25] , indicating that HOTAIR and H3K27me3 might interplay in response to DNA damage. Consist to these findings, the positive correlation of DNA damage with H3K27me3 modification and HOTAIR expression in PBLCs of coke oven workers was revealed in our study.
5. Conclusion
In summary, we found that specific lncRNA HOTAIR and MALAT1 upregulated in PBLCs of the coke oven workers. In addition, the changes of HOTAIR and MALAT1 expression were correlated with both external and internal exposure levels of PAHs and the extent of DNA damage. Our findings reveal that altered HOTAIR and MALAT1 could potentially be used for prediction of PAHs-induced genotoxicity.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by the key program of the Natural Science Foundation of China (NSFC 81430079 , 81130050 ), and NSFC (81402715 , 81273098 ), a National Key Technology Research and Development Program (2014BAI12B02 ), Program of the Guangzhou City Pearl River New Star of Science and Technology (2013J2200020 ), and a program of Natural Science Foundation of Guangdong Province (S2012040007713 ).
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- [1] M.A. Ahmad Chaudhry; Expression pattern of small nucleolar RNA host genes and long non-coding RNA in X-rays-treated lymphoblastoid cells; Int. J. Mol. Sci., 14 (2013), pp. 9099–9110 http://dx.doi.org/10.3390/ijms14059099
- [2] A. Arita, J. Niu, Q. Qu, N. Zhao, Y. Ruan, A. Nadas, Y. Chervona, F. Wu, H. Sun, R.B. Hayes, M. Costa; Global levels of histone modifications in peripheral blood mononuclear cells of subjects with exposure to nickel; Environ. Health Perspect, 120 (2012), pp. 198–203 http://dx.doi.org/10.1289/ehp.1104140
- [3] W. Bai, J. Yang, G. Yang, P. Niu, L. Tian, A. Gao; Long non-coding RNA NR_045623 and NR_028291 involved in benzene hematotoxicity in occupationally benzene-exposed workers; Exp. Mol. Pathol., 96 (2014), pp. 354–360 http://dx.doi.org/10.1016/j.yexmp.2014.02.016
- [4] L. Cantone, F. Nordio, L. Hou, P. Apostoli, M. Bonzini, L. Tarantini, L. Angelici, V. Bollati, A. Zanobetti, J. Schwartz, P.A. Bertazzi, A. Baccarelli; Inhalable metal-rich air particles and histone H3K4 dimethylation and H3K9 acetylation in a cross-sectional study of steel workers; Environ. Health Perspect, 119 (2011), pp. 964–969 http://dx.doi.org/10.1289/ehp.1002955
- [5] J.P. Costantino, C.K. Redmond, A. Bearden; Occupationally related cancer risk among coke oven workers: 30 years of follow-up; J. Occup. Environ. Med., 37 (1995), pp. 597–604 http://dx.doi.org/10.1097/00043764-199505000-00009
- [6] Q. Deng, S. Huang, X. Zhang, W. Zhang, J. Feng, T. Wang, D. Hu, L. Guan, J. Li, X. Dai, H. Deng, X. Zhang, T. Wu; Plasma microRNA expression and micronuclei frequency in workers exposed to polycyclic aromatic hydrocarbons; Environ. Health Perspect, 122 (2014), pp. 719–725 http://dx.doi.org/10.1289/ehp.1307080
- [7] H. Duan, Z. He, J. Ma, B. Zhang, Z. Sheng, P. Bin, J. Cheng, Y. Niu, H. Dong, H. Lin, Y. Dai, B. Zhu, W. Chen, Y. Xiao, Y. Zheng; Global and MGMT promoter hypomethylation independently associated with genomic instability of lymphocytes in subjects exposed to high-dose polycyclic aromatic hydrocarbon; Arch. Toxicol., 87 (2013), pp. 2013–2022 http://dx.doi.org/10.1007/s00204-013-1046-0
- [8] H. Endo, T. Shiroki, T. Nakagawa, M. Yokoyama, K. Tamai, H. Yamanami, T. Fujiya, I. Sato, K. Yamaguchi, N. Tanaka, K. Iijima, T. Shimosegawa, K. Sugamura, K. Satoh; Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer; PLoS One, 8 (2013) http://dx.doi.org/10.1371/journal.pone.0077070
- [9] L. Gao, A. Mai, X. Li, Y. Lai, J. Zheng, Q. Yang, J. Wu, A. Nan, S. Ye, Y. Jiang; LncRNA-DQ786227-mediated cell malignant transformation induced by benzo(a) pyreneNA-DQ786227-mediated cell malignant transformation induced by benzo(a) pyrene; Toxicol. Lett., 223 (2013), pp. 205–210 http://dx.doi.org/10.1016/j.toxlet.2013.09.015
- [10] S. Guil, M. Esteller; RNA -RNA interactions in gene regulation: the coding and noncoding players; Trends Biochem. Sci. (2015), pp. 1–9 http://dx.doi.org/10.1016/j.tibs.2015.03.001
- [11] R.a. Gupta, N. Shah, K.C. Wang, J. Kim, H.M. Horlings, D.J. Wong, M.-C. Tsai, T. Hung, P. Argani, J.L. Rinn, Y. Wang, P. Brzoska, B. Kong, R. Li, R.B. West, M.J. van de Vijver, S. Sukumar, H.Y. Chang; Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis; Nature, 464 (2010), pp. 1071–1076 http://dx.doi.org/10.1038/nature08975
- [12] Y. Han, Y. Liu, Y. Gui, Z. Cai; Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder; J. Surg. Oncol., 107 (2013), pp. 555–559 http://dx.doi.org/10.1002/jso.23264
- [13] G. Hu, T. Yang, J. Zheng, J. Dai, A. Nan, Y. Lai, Y. Zhang, C. Yang, Y. Jiang, E. Lansing; Functional role and mechanism of lncRNA LOC728228 in malignant 16HBE cells transformed by anti-benzopyrene-trans-7,8-dihydrodiol-9,10-epoxide; Mol. Carcinog., 204 (2015), pp. 192–204 http://dx.doi.org/10.1002/mc.22314
- [14] M. Huarte, M. Guttman, D. Feldser, M. Garber, M.J. Koziol, D. Kenzelmann-Broz, A.M. Khalil, O. Zuk, I. Amit, M. Rabani, L.D. Attardi, A. Regev, E.S. Lander, T. Jacks, J.L. Rinn; A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response; Cell, 142 (2010), pp. 409–419 http://dx.doi.org/10.1016/j.cell.2010.06.040
- [15] IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. International Agency for Research on Cancer Iarc Monographs on the Evaluation of Carcinogenic Risks To Humans. Iarc Monogr Eval Carcinog Risks To Humans, 96, i–ix+ 1–390, 2002. doi: 10.1002/food.19940380335.
- [16] I.W.H. Jarvis, K. Dreij, Å. Mattsson, B. Jernström, U. Stenius; Interactions between polycyclic aromatic hydrocarbons in complex mixtures and implications for cancer risk assessment; Toxicology, 321 (2014), pp. 27–39 http://dx.doi.org/10.1016/j.tox.2014.03.012
- [17] P. Ji, S. Diederichs, W. Wang, S. Böing, R. Metzger, P.M. Schneider, N. Tidow, B. Brandt, H. Buerger, E. Bulk, M. Thomas, W.E. Berdel, H. Serve, C. Müller-Tidow; MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer; Oncogene, 22 (2003), pp. 8031–8041 http://dx.doi.org/10.1038/sj.onc.1206928
- [18] D.P. Johnson, G.S. Spitz, S. Tharkar, S.N. Quayle, J.R. Shearstone, S. Jones, M.E. Mcdowell, H. Wellman, J.K. Tyler, B.R. Cairns, M.B. Chandrasekharan; HDAC1, 2 inhibition impairs EZH2- and BBAP- mediated DNA repair to overcome chemoresistance in EZH2 gain-of-function mutant diffuse large B-cell lymphoma; Oncotarget, 6 (2014), pp. 4863–4887
- [19] F.J. Jongeneelen, R.P. Bos, R.B. Anzion, J.L. Theuws, P.T. Henderson; Biological monitoring of polycyclic aromatic hydrocarbons. Metabolites in urine; Scand. J. Work Environ. Health, 12 (1986), pp. 137–143
- [20] M. Kitagawa, K. Kitagawa, Y. Kotake, H. Niida, T. Ohhata; Cell cycle regulation by long non-coding RNAs; Cell. Mol. Life Sci., 70 (2013), pp. 4785–4794 http://dx.doi.org/10.1007/s00018-013-1423-0
- [21] R. Kogo, T. Shimamura, K. Mimori, K. Kawahara, S. Imoto, T. Sudo, F. Tanaka, K. Shibata, A. Suzuki, S. Komune, S. Miyano, M. Mori; Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers; Cancer Res., 71 (2011), pp. 6320–6326 http://dx.doi.org/10.1158/0008-5472.can-11-1021
- [22] J. Krell, A.E. Frampton, R. Mirnezami, V. Harding, A. De Giorgio, L.R. Alonso, P. Cohen, S. Ottaviani, T. Colombo, J. Jacob, L. Pellegrino, G. Buchanan, J. Stebbing, L. Castellano; Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer; PLoS One, 9 (2014) http://dx.doi.org/10.1371/journal.pone.0098561
- [24] D. Li, Q. Wang, C. Liu, H. Duan, X. Zeng, B. Zhang, X. Li, J. Zhao, S. Tang, Z. Li, X. Xing, P. Yang, L. Chen, J. Zeng, X. Zhu, S. Zhang, Z. Zhang, L. Ma, Z. He, E. Wang, Y. Xiao, Y. Zheng, W. Chen; Aberrant expression of miR-638 contributes to benzo(a) pyrene-induced human cell transformationrant expression of miR-638 contributes to benzo(a) pyrene-induced human cell transformation; Toxicol. Sci., 125 (2012), pp. 382–391 http://dx.doi.org/10.1093/toxsci/kfr299
- [25] Z. Li, H. Mon, H. Mitsunobu, L. Zhu, J. Xu, J.M. Lee, T. Kusakabe; Dynamics of polycomb proteins-mediated histone modifications during UV irradiation-induced DNA damage; Insect Biochem. Mol. Biol., 55 (2014), pp. 9–18 http://dx.doi.org/10.1016/j.ibmb.2014.10.001
- [26] Q. Liu, S. Sun, W. Yu, J. Jiang, F. Zhuo, G. Qiu, S. Xu, X. Jiang; Altered expression of long non-coding RNAs during genotoxic stress-induced cell death in human glioma cells; J. Neurooncol. (2015), pp. 283–292 http://dx.doi.org/10.1007/s11060-015-1718-0
- [27] Y. Liu, F. Luo, Y. Xu, B. Wang, Y. Zhao, W. Xu, L. Shi, X. Lu, Q. Liu; Epithelial-mesenchymal transition and cancer stem cells, mediated by a long non-coding RNA, HOTAIR, are involved in cell malignant transformation induced by cigarette smoke extract; Toxicol. Appl. Pharmacol., 282 (2015), pp. 9–19 http://dx.doi.org/10.1016/j.taap.2014.10.022
- [28] Y. Liu, S. Yang, X. Zhang; Down-regulation of long non-coding RNA TUG1 suppresses melanoma cell proliferation and induces apoptosis via up-regulating microRNA-9; Biochem. Biophys. Res. Commun. (2013) http://dx.doi.org/10.1016/j.bbrc.2013.09.050 65138576
- [29] A.K. Marrone, F.A. Beland, I.P. Pogribny; Noncoding RNA response to xenobiotic exposure: an indicator of toxicity and carcinogenicity; Expert Opin. Drug Metab. Toxicol., 10 (2014), pp. 1409–1422
- [30] M. Mourtada-Maarabouni, M.R. Pickard, V.L. Hedge, F. Farzaneh, G.T. Williams; GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer; Oncogene, 28 (2009), pp. 195–208 http://dx.doi.org/10.1038/onc.2008.373
- [31] E. Özgür, U. Mert, M. Isin, M. Okutan, N. Dalay, U. Gezer; Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells; Clin. Exp. Med., 13 (2013), pp. 119–126 http://dx.doi.org/10.1007/s10238-012-0181-x
- [32] S. Pavanello, V. Bollati, A.C. Pesatori, L. Kapka, C. Bolognesi, P.A. Bertazzi, A. Baccarelli; Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals; Int. J. Cancer, 125 (2009), pp. 1692–1697 http://dx.doi.org/10.1002/ijc.24492
- [33] V.J. Peschansky, C. Wahlestedt; Non-coding RNAs as direct and indirect modulators of epigenetic regulation; Epigenetics, 9 (2014), pp. 3–12 http://dx.doi.org/10.4161/epi.27473
- [34] X. Shi, M. Sun, H. Liu, Y. Yao, R. Kong, F. Chen, Y. Song; A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer; Mol. Carcinog., 12 (1–12) (2013) http://dx.doi.org/10.1002/mc.22120
- [35] N.P. Singh, M.T. McCoy, R.R. Tice, E.L. Schneider; A simple technique for quantitation of low levels of DNA damage in individual cells; Exp. Cell Res., 175 (1988), pp. 184–191 90265-0 http://dx.doi.org/10.1016/0014-4827(88) 90265-0
- [36] R.a. Stein; Epigenetics and environmental exposures; J. Epidemiol. Community Health, 66 (2012), pp. 8–13 http://dx.doi.org/10.1136/jech.2010.130690
- [37] L. Tang, W. Zhang, B. Su, B. Yu; Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma; Biomed. Res. Int., 2013 (2013), p. 251098
- [38] Y. Tian, X. Zhang, Y. Hao, Z. Fang, Y. He; Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma; Melanoma Res. (2014) http://dx.doi.org/10.1097/cmr.0000000000000080
- [39] V. Tripathi, Z. Shen, A. Chakraborty, S. Giri, S.M. Freier, X. Wu, Y. Zhang, M. Gorospe, S.G. Prasanth, A. Lal, K.V. Prasanth; Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB; PLoS Genet., 9 (2013) http://dx.doi.org/10.1371/journal.pgen.1003368
- [40] F. Wang, S. Ren, R. Chen, J. Lu, X. Shi, Y. Zhu; Development and prospective multicenter evaluation of the long noncoding RNA MA LAT-1 as a diagnostic urinary biomarker for prostate cancer; Oncotarget, 5 (2014)
- [41] L. Wang, X. Zeng, S. Chen, L. Ding, J. Zhong, J.C. Zhao, L. Wang, A. Sarver, A. Koller, J. Zhi, Y. Ma, J. Yu, J. Chen, H. Huang; BRCA1 is a negative modulator of the PRC2 complex; EMBO J., 32 (2013), pp. 1584–1597 http://dx.doi.org/10.1038/emboj.2013.95
- [42] Y. Wei, B. Niu; Role of MALAT1 as a prognostic factor for survival in various cancers: a systematic review of the literature with meta-analysis; Dis. Markers, 2015 (2015), pp. 1–9 http://dx.doi.org/10.1155/2015/164635
- [43] Y. Wu, L. Zhang, Y. Wang, H. Li, X. Ren, F. Wei, W. Yu, X. Wang, L. Zhang, J. Yu, X. Hao; Long noncoding RNA HOTAIR involvement in cancer; Tumor Biol., 35 (2014), pp. 9531–9538 http://dx.doi.org/10.1007/s13277-014-2523-7
- [44] Z.-Y. Xu, Q.-M. Yu, Y.-A. Du, L.-T. Yang, R.-Z. Dong, L. Huang, P.-F. Yu, X.-D. Cheng; Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer; Int. J. Biol. Sci., 9 (2013), pp. 587–597 http://dx.doi.org/10.7150/ijbs.6339
- [45] G. Yang, X. Lu, L. Yuan; LncRNA: a link between RNA and cancer; Biochim. Biophys. Acta- Gene Regul. Mech., 1839 (11) (2014), pp. 1097–1109 http://dx.doi.org/10.1016/j.bbagrm.2014.08.012
- [46] P. Yang, J. Ma, B. Zhang, H. Duan, Z. He, J. Zeng, X. Zeng, D. Li, Q. Wang, Y. Xiao, C. Liu, Q. Xiao, L. Chen, X. Zhu, X. Xing, Z. Li, S. Zhang, Z. Zhang, L. Ma, E. Wang, Z. Zhuang, Y. Zheng, W. Chen; CpG site-specific hypermethylation of p16 INK4α in peripheral blood lymphocytes of PAH-exposed workers; Cancer Epidemiol. Biomarkers Prev., 21 (2012), pp. 182–190 http://dx.doi.org/10.1158/1055-9965.epi-11-0784
- [47] Q. Yang, S. Zhang, H. Liu, J. Wu, E. Xu, B. Peng, Y. Jiang; Oncogenic role of long noncoding RNA AF118081 in anti-benzo[a]pyrene-trans-7,8-dihydrodiol-9,10-epoxide-transformed 16HBE cells; Toxicol. Lett., 229 (2014), pp. 430–439 http://dx.doi.org/10.1016/j.toxlet.2014.07.004
- [48] A. Zhang, M. Xu, Y.Y. Mo; Role of the lncRNA-p53 regulatory network in cancer; J. Mol. Cell Biol. (2014) http://dx.doi.org/10.1093/jmcb/mju013
- [49] E. Zhang, D. Yin, M. Sun, R. Kong, X. Liu, L. You, L. Han, R. Xia, K. Wang, J. Yang, W. De, Y. Shu, Z. Wang; P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression; Cell Death Dis., 5 (2014), p. e1243 http://dx.doi.org/10.1038/cddis.2014.201
- [50] Q. Zhang, P.L. Geng, P. Yin, X.L. Wang, J.P. Jia, J. Yao; Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis; Asian Pac. J. Cancer Prev., 14 (2013), pp. 2311–2315 http://dx.doi.org/10.7314/apjcp.2013.14.4.2311
- [51] Y. Zhang, S. Tao, H. Shen, J. Ma; Inhalation exposure to ambient polycyclic aromatic hydrocarbons and lung cancer risk of Chinese population; Proc. Natl. Acad. Sci. U. S. A., 106 (2009), pp. 21063–21067 http://dx.doi.org/10.1073/pnas.0905756106
Document information
Published on 02/05/17
Accepted on 02/05/17
Submitted on 02/05/17
Licence: Other
Share this document
Keywords
claim authorship
Are you one of the authors of this document?