Highlights
- Functional antibodies to influenza matrix 1 and nucleoprotein are common in healthy and influenza-infected humans.
- Opsonising antibodies to matrix 1 and nucleoprotein can bind FcγRIIIa dimers and activate natural killer cells.
- Influenza infection increased natural killer cell activation to hemagglutinin but changes to the internal proteins varied
Influenza virus causes both seasonal outbreaks and global pandemics. The current influenza vaccine provides minimal protection against divergent strains of the virus not found in the vaccine. While neutralising antibodies induced by vaccination are able to confer strain-specific protection, antibodies directed against conserved influenza proteins may be able to provide some cross-protection. Animal models suggest a protective role for anti-nucleoprotein antibodies. Exploring the functional capacity of human antibodies against internal influenza proteins to engage Fc receptors and activate innate immune cells may present a unique approach in the development of a more universal influenza vaccine.
Abstract
The conserved internal influenza proteins nucleoprotein (NP) and matrix 1 (M1) are well characterised for T cell immunity, but whether they also elicit functional antibodies capable of activating natural killer (NK) cells has not been explored. We studied NP and M1-specific ADCC activity using biochemical, NK cell activation and killing assays with plasma from healthy and influenza-infected subjects. Healthy adults had antibodies to M1 and NP capable of binding dimeric FcγRIIIa and activating NK cells. Natural symptomatic and experimental influenza infections resulted in a rise in antibody dependent NK cell activation post-infection to the hemagglutinin of the infecting strain, but changes in NK cell activation to M1 and NP were variable. Although antibody dependent killing of target cells infected with vaccinia viruses expressing internal influenza proteins was not detected, opsonising antibodies to NP and M1 likely contribute to an antiviral microenvironment by stimulating innate immune cells to secrete cytokines early in infection. We conclude that effector cell activating antibodies to conserved internal influenza proteins are common in healthy and influenza-infected adults. Given the significance of such antibodies in animal models of heterologous influenza infection, the definition of their importance and mechanism of action in human immunity to influenza is essential.
Keywords
Influenza ; Natural killer cells ; Antibody dependent cellular cytotoxicity ; Nucleoprotein ; Matrix protein 1 ; Hemagglutinin
1. Introduction
Seasonal influenza epidemics result in 30 to 50 million cases of severe illness and 250 to 500 thousand deaths each year mainly in the very young, the elderly and the chronically ill. To prevent influenza infection a trivalent or quadrivalent vaccine, containing inactivated split virion preparations from two influenza A viruses (H1N1 and H3N2) and one or two influenza B viruses, is widely administered especially to high-risk groups (World Health Organization, 2015 ). Influenza vaccination primarily induces neutralising antibodies (NAbs) against the envelope protein hemagglutinin (HA) (Atmar et al., 2007 and Martin Mdel et al., 2010 ). NAbs bind to HA inhibiting viral attachment to cell surface sialic acid and prevent viral fusion with host cells (Krause et al., 2011 and Wang et al., 2010 ). However the effectiveness of antibody-mediated neutralisation is limited by the continual antigenic drift of the HA protein (Sandbulte et al., 2011 and Shil et al., 2011 ). The accumulation of mutations in HA and re-assortment events between antigenically distinct viruses (antigenic shift) can result in the emergence of influenza variants not recognized by NAbs (Fielding et al., 2011 , Hardelid et al., 2011 and Yang et al., 2012 ). To minimize the impact of antigenic change the influenza vaccine is updated annually. Furthermore seasonal vaccination is strain-specific and is only weakly protective against infection with divergent influenza viruses. Global influenza pandemics can occur when there is minimal immunity to novel influenza viruses capable of infecting humans (Peiris et al., 2009 ).
In addition to neutralisation, antibodies (Abs) mediate a number of other effector functions through their Fc region. These functions include complement activation (O'Brien et al., 2011 and Ohta et al., 2011 ), Ab-dependent phagocytosis (ADP) (Huber et al., 2001 ) and Ab-dependent cellular cytotoxicity (ADCC) (Greenberg et al., 1978 , Hashimoto et al., 1983a , Hashimoto et al., 1983b and Vella et al., 1980 ). Natural killer (NK) cells are cytotoxic lymphocytes of the innate immune system that play an important role in the elimination of virally infected and transformed cells. ADCC is mediated when the NK cell FcγRIIIa (CD16a) receptor interacts with the Fc region of IgG Abs bound to their viral antigens (Ags) on the surface of virally infected cells. IgG-Fc region binding to and crosslinking of FcγRIIIa leads to NK cell activation, which includes degranulation releasing perforins and granzymes, as well as the secretion of anti-viral cytokines such as IFNγ and TNF. It has been shown that ADCC Abs enhance protection against several clinically important viral pathogens (Laoprasopwattana et al., 2007 , Vogt et al., 2011 and Xiao et al., 2010 ). Our group has previously demonstrated that healthy influenza-exposed adults have Abs to HAs from non-circulating strains of influenza in the absence of NAbs. We have also shown that HA-specific Abs lead to NK cell activation and influenza clearance in vitro (Jegaskanda et al., 2013a , Jegaskanda et al., 2013b , Jegaskanda et al., 2013c and Jegaskanda et al., 2014a ). There has been a recent interest in ADCC Abs targeting more conserved influenza surface Ags such as the extracellular domain of matrix 2 protein (eM2) (El Bakkouri et al., 2011 , Kim et al., 2014 and Lee et al., 2014 ) and the stalk region of HA (DiLillo et al., 2014 , Ellebedy et al., 2014 , Margine et al., 2013a and Margine et al., 2013b ).
Influenza proteins inside the viral envelope are highly conserved across different strains and subtypes of influenza. Two internal proteins that have been extensively studied in the context of influenza infection are nucleoprotein (NP) and matrix protein 1 (M1). NP binds and transports the RNA genome of influenza and M1 is a structural protein that coats the inside of the influenza virion. Both NP and M1 contain well-characterized epitopes for CD4+ and CD8+ T-cell responses to influenza virus infection. Ongoing studies aim to create a heterosubtypic influenza vaccine by specifically inducing T-cell responses to conserved NP and M1 epitopes (Chen et al., 2014 , Hessel et al., 2014 , Lambe et al., 2013 and Lillie et al., 2012 ). An additional role for Abs against internal influenza proteins is starting to emerge. In animal models, NP-specific IgG and vaccination with whole NP have successfully provided protection against heterologous influenza challenge (Epstein et al., 2005 , Lamere et al., 2011a , Ulmer et al., 1993 and Wraith et al., 1987 ). Passive transfer of non-neutralising NP Abs can also fully protect against influenza challenge in mice, but the mechanism of protection has yet to be elucidated (Carragher et al., 2008 and LaMere et al., 2011b ). NP can also be found on the surface of influenza-infected cells in vitro, along with barely detectable levels of M1 (Bodewes et al., 2013 , Virelizier et al., 1977 and Yewdell et al., 1981 ). It is conceivable that non-neutralising Abs to M1 and NP may be involved in clearance of infected cells through Fc mediated effector functions. Improving our understanding of the humoral response to internal influenza proteins may contribute to the development of a universally protective vaccination strategy. Herein we have demonstrated M1 and NP Abs are a common feature of influenza-exposed or infected human adults and these Abs have the ability to trigger activation of NK cells.
2. Materials and Methods
2.1. Donors and Plasma Samples
We studied healthy adults, subjects with clinical influenza virus infection and subjects with experimental influenza virus infection for NK cell activating Abs to internal influenza proteins. Fourteen healthy donors were recruited to donate plasma samples from September 2013 to December 2014. The median age of the donors is 29.5, with a range of 22–52. All of the donors had the trivalent influenza vaccine one to 10 times since 2005. Plasma samples were also taken from four influenza-naïve pigtail macaques (Macaca nemestrina ) to serve as influenza Ab negative samples. Nine of the healthy donors recruited above and one of the influenza-naïve macaques were used in the recombinant soluble FcγRIIIa ELISA described below. Plasma samples from two of the healthy donors and one macaque were also tested in an NK cell activation assay with purified primary NK cells. Macaque studies were approved by the Commonwealth Scientific and Industrial Research Organizational animal health ethics committee. The WHO Collaborating Centre for Reference and Research on Influenza provided six sera samples from three subjects collected in 2013 with likely A/California/07/2009 (H1N1 pdm09) influenza virus infection. One serum sample was taken at the patients initial doctors visit with an influenza-like illness and a second sample was collected approximately two weeks later at a follow-up appointment. Influenza H1N1 pdm09 virus infection was confirmed by a complement fixation test (CFT) and hemagglutination inhibition (HI) assay ( Table 1 ). Plasma samples from 11 volunteers challenged with A/Wisconsin/67/2005 (H3N2) influenza were provided from a previously published study conducted on behalf of the University of Oxford. Initially all participants, aged 18–45, had undetectable serum Abs to the challenge A/Wisconsin/67/2005 (H3N2) strain (HI ≤ 10). The volunteers were infected with A/Wisconsin/67/2005 (H3N2) intranasally while under quarantine. They were monitored for viral shedding, disease symptoms and a HI assay was performed 36 days post-challenge (Table 2 ) (Lillie et al., 2012 ). Healthy blood donors provided PBMCs as a source of NK cells in some assays. The relevant human ethics committees approved all studies.
| Patient | Time point | CFT result | HI titre H1N1 A/California/07/2009 |
|---|---|---|---|
| 1 | Day 0 | < 10 | < 10 |
| Day 17 | > 120 | 80 | |
| 2 | Day 0 | 10 | < 10 |
| Day 16 | > 320 | 20 | |
| 3 | Day 0 | 10 | < 10 |
| Day 12 | > 320 | 80 |
| Subject | Disease severity | Virus shed | HI titre to A/Wisconsin/67/2005 | Percent change of CD107a+ NK-92s to H3 Wsn05 protein post-infection | Percent change CD107a+ NK-92s to M1 protein post-infection | Percent change CD107a+ NK-92s to NP protein post-infection |
|---|---|---|---|---|---|---|
| 72 | Moderate/severe | N | 80 | 91.2 | 80.3 | 26.5 |
| 84 | Moderate/severe | Y | 40 | 21.1 | 0.7 | 2.1 |
| 87 | Moderate/severe | Y | 80 | 52.8 | − 22.8 | − 0.7 |
| 100 | Moderate/severe | Y | 160 | 23.1 | − 14.9 | 6.5 |
| 81 | Mild | Y | < 10 | 0.4 | − 7.8 | − 1.3 |
| 95 | Mild | Y | < 10 | − 5.7 | 8.8 | 28.9 |
| 96 | Mild | N | 20 | − 6.8 | − 4.6 | − 1.4 |
| 109 | Mild | N | 80 | 10.4 | 9.0 | 3.7 |
| 86 | None | N | 320 | 12.4 | 22.7 | 21.3 |
| 93 | None | N | 320 | 3.4 | 3.4 | − 4.3 |
| 108 | None | N | 20 | 23.5 | 17.6 | 10.5 |
| Median percent change of CD107a+ NK-92s post-infection | 12.4 | 3.4 | 3.7 | |||
⁎ Table adapted from previously published work by Lillie et al. (2012)
2.2. Intravenous Immunoglobulin (IVIG)
We studied 18 IVIG preparations collected over 7 years from 2004 to 2010 as a representation of pooled IgG from thousands of healthy HIV negative donors (Sandoglobulin, CSL Behring, Australia). The 18 IVIG preparations were made by cold ethanol fractionation and were 96% IgG. Stock IVIG preparations are at a concentration of 60-200 mg/ml and prior to use all preparations were diluted to a concentration of 10 mg/ml in Fetal Calf Serum (FCS) as previously described (Jegaskanda et al., 2014a ).
2.3. Cell Lines and Viruses
The NK cell line GFP-CD16 (176V) NK-92 (Gong et al., 1994 ) was used to perform some of the NK cell activation assays. The parental cells of GFP-CD16 (176V) NK-92 are NK-92 cells from the ATCC (CRL-2407). GFP-CD16 (176V) NK-92 cells have been transduced with a retrovirus to express the high affinity variant of CD16 (V176) in the pBMN-IRES-EGFP vector. On these cells surface expression of CD16 (V176) correlates with GFP expression. GFP-CD16 (176V) NK-92 cells were kindly provided by Dr. Kerry Campbell from the Institute for Cancer Research in Philadelphia, PA.
A549 (ATCC) and CEM.NKr-CCR5 cells (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH) were used as targets for cytotoxicity assays. Recombinant vaccinia viruses (rVV) were generated by homologous recombination of vaccinia infected cells with non-conjugative plasmids expressing influenza genes into the vaccinia 7.5 k or TK genes as previously described (Mackett et al., 1984 ). rVVs containing individual HA, NP or M1 genes from influenza A/Puerto Rico/8/1934 (H1N1) were kindly provided by Drs. Jonathan Yewdell and Jack Bennink (NIH, Bethesda, MD) and expanded by Dr. Weisan Chen (LaTrobe University, Melbourne, VIC) (Smith et al., 1987 and Yewdell et al., 1985 ).
2.4. NK Cell Activation Assay
NK cell activation assays performed with influenza-infected cells as targets and polyclonal serum does not allow for the study of responses to individual Influenza proteins. Thus, NK cell activation was measured by the ability of Abs bound to a plate coated with influenza proteins to induce NK cell expression of IFNγ and CD107a as previously described (Jegaskanda et al., 2013a , Jegaskanda et al., 2013b , Jegaskanda et al., 2013c , Jegaskanda et al., 2013d , Jegaskanda et al., 2014a and Jegaskanda et al., 2014b ). Briefly, 96-well ELISA plates (Nunc, Rochester, NY) were coated with 600 ng of purified influenza protein (Sino Biological, Shanghai, China) overnight at 4 °C in PBS. The wells were washed five times with PBS and incubated with heat-inactivated (56 °C for 1 h) undiluted sera/plasma or IVIG for 2 h at 37 °C. Plates were washed seven times with PBS and 106 peripheral blood mononuclear cells (PBMCs) or 5 × 105 purified NK cells were added to each well. Healthy donor PBMCs were obtained from buffy packs provided by the Australian Red Cross. PBMCs were isolated by Ficoll-Paque PLUS (GE Healthcare, Madison, WI), washed with RF10 (RPMI 1640 supplemented with 10% FCS, penicillin, streptomycin and l-glutamine; Life Technologies, Grand Island, NY), frozen in FCS containing 10% DMSO and stored in liquid nitrogen. Once thawed PBMCs were washed twice with RF10 before addition to each well. Purified NK cells were made from fresh PBMCs using the EasySep Human NK Cell Enrichment Kit (Stemcell Technologies, Vancouver, Canada). Anti-human CD107a allophycocyanin-H7 Ab (clone H4B4; BD Biosciences, San Jose, CA), 5 μg/ml brefeldin A (Sigma Aldrich) and 5 μg/ml monensin (Golgi Stop; BD Biosciences) were added to the cells and incubated for 5 h at 37 °C with 5% CO2 . PBMCs were then incubated with 1 mM EDTA to minimize cell adherence to the plates, anti-human CD3 PerCP (clone SP34-2) and anti-human CD56 allophycocyanin (clone B159; both from BD Biosciences) for 30 min at room temperature in the dark. Cells were fixed with 1% formaldehyde (Sigma Aldrich, St. Louis, M1) for 10 min and permeabilized with FACS permeabilizing solution 2 (BD Biosciences) for 10 min. PBMCs were then incubated at room temperature for 1 h with IFNγ AF700 (clone B27; BD Biosciences) in the dark. Finally, cells were again fixed with 1% formaldehyde and 50,000–100,000 (20,000–50,000 for purified NK cells) events were generally acquired on the LSRII flow cytometer (BD Biosciences). Plasma from influenza-naïve pigtail macaques was used as a negative control in some NK cell activation assays. Virtually all humans are serially exposed to influenza viruses from early childhood and influenza-naïve humans are rare. We have previously shown that Ig purified from influenza-exposed pigtail macaques can induce activation of primary human NK cells, whereas naïve macaque plasma collected prior to influenza exposure failed to activate NK cells (Jegaskanda et al., 2013a ).
GFP-CD16 (176V) NK-92 cells and CD16-negative parental NK-92 cells were also used in some NK cell activation assays that required a large number of cells. The ELISA plate coating was performed as described above. Following PBS washing, however, an additional blocking step was performed with PBS containing 5% Bovine serum albumin (Sigma Aldrich, St. Louis, MI) and 0.1% Tween 20 (U-CyTech) for 2 h at 37 °C. Once blocked, plates were washed with PBS and incubated with heat-inactivated sera/plasma or IVIG for 2 h at 37 °C. Plates were washed with PBS, 2 × 105 GFP-CD16 (176V) NK-92 cells were added to each well and incubated at 37 °C with 5% CO2 for 5 h. Anti-human CD107a allophycocyanin (clone H4A3; BD Biosciences) and 1 mM EDTA were added to the cells for 30 min at room temperature in the dark. GFP-CD16 (176 V) NK-92 cells were then washed twice with PBS, fixed with 1% formaldehyde and 50,000–100,000 events were generally acquired on the LSRII flow cytometer. Analysis was performed using FlowJo X 10.0.7r2 software (FlowJo LLC, Ashland, OR).
2.5. Dimeric Recombinant Soluble FcγRIIIa (CD16a) Binding ELISA
We recently developed a dimeric recombinant soluble FcγRIIIa (rsFcγRIIIa) ELISA to model the need for ADCC-inducing Abs to cross link FcγRIIIa (unpublished data; Wines et al). For the dimeric rsFcγRIIIa binding assay, a 96-well ELISA plate was coated with 50 ng of purified influenza protein or PBS overnight at 4 °C in PBS. The plate was washed twice with PBST and blocked with 140 μl of PBS 1 mM EDTA 1% BSA (PBSE/BSA) for 1 h at 37 °C. The plate was then washed twice with PBST and 50 μl of half log plasma dilutions (1:10, 1:32, 1:100 and 1:320) in PBSE/BSA was added. Following addition of plasma dilutions, the plate was incubated at 37 °C for 1 h. The ELISA plate was washed five times with PBST and 50 μl of 0.1 μg/ml rsFcγRIIIa (V176) dimer was added to the wells then the plate was incubated for 1 h at 37 °C. Pierce High Sensitivity HRP-Streptavidin (Thermo Fisher Scientific, Waltham, MA) was diluted 1:10,000 in PBSE/BSA, added to all wells and the plate was incubated at 37 °C for 1 h. The plate was washed eight times with PBST and blotted dry. 50 μl of TMB substrate was added to each well and the plate was developed for 4–8 min in the dark. The reaction was stopped with 1 M HCl and the plate was read at an absorbance of 450 nm. Intragam 5 (bioCSL, Melbourne, VIC) diluted in PBSE to 5 μg/ml was coated on all plates as a positive control and allowed for normalisation between plates in large assays.
2.6. ADCC Killing Assay With Recombinant Vaccinia Viruses (rVVs) Expressing Single Influenza Proteins
To study ADCC-mediated cytotoxicity, we adapted an ADCC assay that measures LDH release from target cells as previously described (Cox, 1999 and Gooneratne et al., 2015 ). Briefly, CEM.NKr-CCR5 cells or A549 cells were infected with rVVs (either wild type rVV, rVV NP, rVV M1 or rVV HA, kindly provided by Dr J. Yewdell and Dr J. Bennink, NIH) in 0.1% BSA PBS at an MOI of 10 for one hour at 37 °C. Following infection the wells were topped up with RF10 and incubated for 8 h or 20 h at 37 °C with 5% CO2 . The rVV-infected cells were then washed three times with fresh RF10 media. The CytoTox 96 nonradioactive cytotoxicity kit (Promega, Madison, WI) was utilized to measure lactate dehydrogenase (LDH) release from the cytoplasm of cells killed by ADCC. The rVV-infected cells were the targets with freshly isolated healthy donor PBMCs or NK-92 cells as the effectors. Spontaneous release of LDH from target and effector cells was determined by incubation of each cell type alone. Maximum LDH release from target cells was determined by addition of lysis buffer to target cells during the 4 h incubation at 37 °C. Background levels of LDH in RF10 media were determined by media only control wells. In assays were CEMs were used as targets, experimental wells contained 2 × 104 rVV-infected CEM cells combined with 5 × 105 or 2.5 × 105 PBMCs in an effector to target ratio of 25:1 or 12.5:1. When A549 cells were used as targets, experimental wells contained 3 × 104 rVV-infected A549 cells combined with 7.5 × 105 PBMCs in an effector to target ratio of 25:1. In assays where NK-92 cells were used as effectors, experimental wells contained 2 × 104 rVV-infected target cells combined with 1 × 105 or 2 × 104 NK-92s in an effector to target ratio of 5:1 or 1:1 respectively. To measure influenza-specific ADCC, IgG was purified from a healthy influenza-exposed adult, with known M1 and NP-specific NK cell activating Abs and who was not immunised against VV, using the Protein G HP Multitrap and Antibody Buffer Kit (both GE Healthcare, UK) according to the manufacturers instructions. Purified IgG was added to the experimental wells giving final concentrations of 50 μg/ml, 5 μg/ml and 0.5 μg/ml. Killing of infected target cells by effectors in the absence of Ab was measured by adding PBS to wells containing both the effectors and targets (no Ab control). We assessed ADCC to target cells infected with five different rVVs, therefore experimental and control conditions were prepared separately for target cells infected with each virus. Additionally all control and all experimental conditions were set-up in triplicate. Once all reagents were added to the appropriate wells the plates were spun at 250 × g for four minutes then incubated at 37 °C 5% CO2 for 4 h. Following incubation, the plates were again spun at 250 × g for 4 min then 50 μl of supernatant was transferred to another flat-bottom 96-well plate. 50 μl of substrate solution was added to wells containing supernatant and plates were incubated at room temperature in the dark for 30 min. The reaction was then stopped with 50 μl of stop solution and the absorbance was recorded at 490 nm. The optical density of the media only control was subtracted from all other values. The following formula was then used to calculate percentage cytotoxicity for all experimental conditions % cytotoxicity = [(experimental − effector spontaneous − target spontaneous) / (maximum LDH − target spontaneous)].
2.7. Statistical Analysis
Statistical analysis was performed with Prism GraphPad version 5.0d (GraphPad Software, San Diego, CA). Data presented in Fig. 1 b and c were analysed by Mann Whitney U test to compare NK cell activation by plasma from influenza-exposed humans to NK cell activation by plasma from influenza-naïve macaques. A Friedman test was used to determine if there was a significant overall difference in NK cell activation for the same set of samples (14 healthy donors in Fig. 1 b, c; 18 IVIG preparations in Fig. 4 a, b) exposed to multiple conditions (HA vs M1 vs NP vs gp140). A Wilcoxon matched pairs signed-rank test was used, alone or in concert with a Friedman test, to pinpoint whether there was a significant difference in NK cell activation for paired samples exposed to two separate conditions (influenza protein vs irrelevant HIV-1 protein for Figs. 1 b, c and 4 a, b; pre- vs post-infection for Figs. 6 a–c and 7 ). The Wilcoxon matched pairs signed-rank test was sometimes performed multiple times on the same data set therefore a Bonferroni correction was used to correct the p value for multiple comparisons (Fig. 1 b, c; Fig. 4 a, b). A nonparametric Spearmen correlation was performed to determine whether there was a statistically significant correlation between two data sets (Fig. 2 c, e; Fig. 3 b, c; Fig. 5 c; Fig. 6 d).
|
|
|
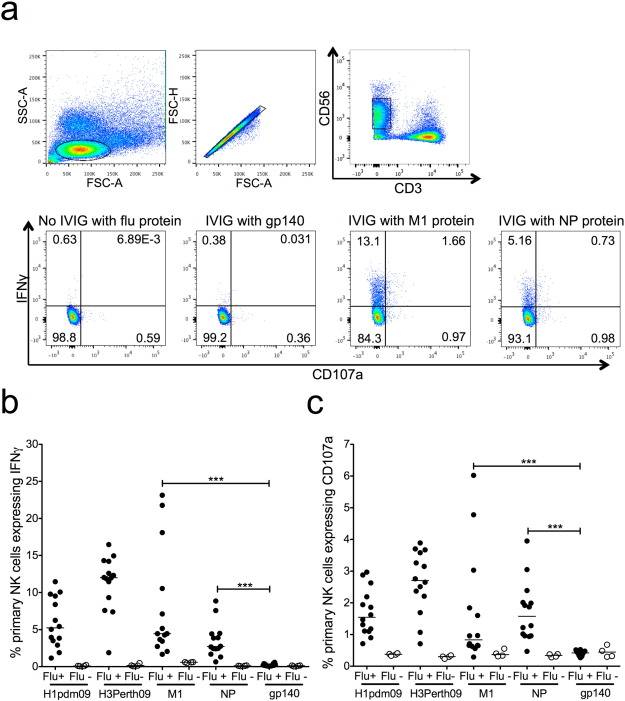
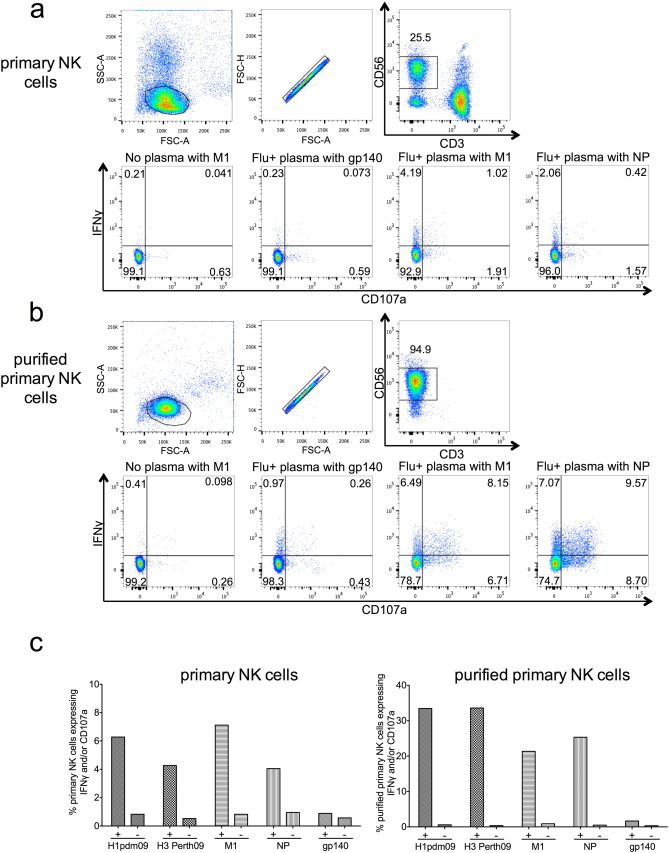
Fig. 1. M1- and NP-specific primary NK cell activation in healthy influenza-exposed adults. a) Lymphocytes were gated on by size and granularity (FSC-A vs SSC-A) ensuring single cells (FSC-A vs FSC-H). CD3 − CD56 + dim primary NK cells were selected for analysis using IFNγ and CD107a as activation markers. PBMCs were incubated with influenza protein (600 ng/well) in the absence of IVIG from influenza-exposed adults, irrelevant viral protein gp140 (600 ng/well) with IVIG from influenza-exposed adults and influenza proteins (M1 and NP) with IVIG from influenza-exposed adults. Primary NK cell activation with plasma from 14 healthy adults (Flu +) and four influenza-naïve pigtail macaques (Flu −) is shown by IFNγ (b) and CD107a (c) expression to HA of A/California/04/2009 (H1pdm09), HA of A/Perth/19/2009 (H3Perth09), M1 of A/Puerto Rico/8/1934 (M1), NP of A/California/07/2009 (NP) and irrelevant viral protein gp140. Values are unsubtracted with gp140 background shown for all samples. For each influenza protein tested Flu + and Flu − groups were compared with a Mann Whitney U test where p < 0.05 was considered significant. A Friedman test followed by a Wilcoxon matched pairs signed-rank test with a Bonferroni correction for multiple comparisons was used to compare the Flu + group incubated with M1 and NP to the Flu + group incubated with irrelevant protein gp140, a corrected p value of < 0.0125 considered significant. *** = p < 0.001. |
|
|
|
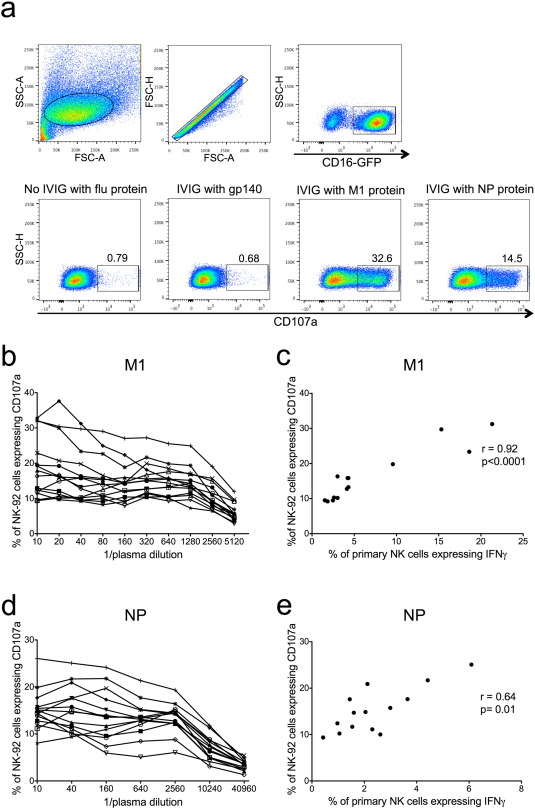
Fig. 2. Titration of M1- and NP-specific NK activating Abs in healthy adults with NK-92 cells and correlation with primary NK cells. a) NK-92 cells were gated on by size and granularity (FSC-A vs SSC-A) ensuring single cells. CD16-GFP + cells were selected for analysis using CD107a as an activation marker. NK-92 cells were incubated with influenza protein (600 ng/well) in the absence of IVIG from influenza-exposed adults, irrelevant viral protein gp140 (600 ng/well) with IVIG from influenza-exposed adults and influenza proteins (M1 and NP) with IVIG from influenza-exposed adults. 14 healthy donor plasma samples previously screened for primary NK cell activation in the presence of influenza proteins (Fig. 1 b, c) were titrated for NK cell activating Abs to M1 (b) in a series of 2-fold plasma dilutions and NP (d) in a series of 4-fold plasma dilutions. All values were background subtracted with wells containing plasma and the irrelevant HIV-1 protein gp140. Correlations between primary NK cell activation (percentage of NK cells expressing IFNγ) with undiluted plasma and NK-92 activation (percentage of NK-92 cells expressing CD107a) with a 1:40 plasma dilution are shown for M1 (c) and NP (e). Spearman correlation was used to determine correlation between primary NK and NK-92 cell activation where p < 0.05 was considered significant. |
|
|
|
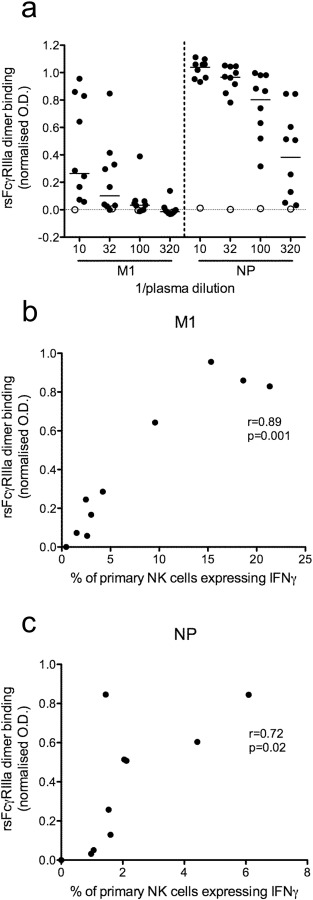
Fig. 3. NP and M1 opsonised with Abs from healthy influenza-exposed adults bind dimeric rsFcγRIIIa. a) A subset of 9 healthy human donors (closed circle) and one influenza-naïve macaque (open circle) previously screened for NK cell activating Abs to M1 and NP (Fig. 1 and Fig. 2 ) were tested for dimeric rsFcγRIIIa binding to Ab opsonised M1 and NP using a novel ELISA. Three half-log plasma dilutions starting at 1:10 were screened for each donor. All normalised OD values were subtracted with a no antigen control well, containing plasma but no influenza protein. Correlations between primary NK cell activation (percentage of NK cells expressing IFNγ) and dimeric rsFcγRIIIa binding with a plasma dilution of 1:10 for M1 (b) and 1:320 for NP (c) are shown. All percentages of primary NK cells expressing IFNγ are background subtracted with wells containing plasma and the irrelevant HIV-1 protein gp140. Spearman correlation was used to determine correlation between primary NK cell activation and dimeric rsFcγRIIIa binding where p < 0.05 was considered significant. |
2.8. Role of the Funding Source
Funding was generously provided by the Australian National Health and Medical Research Council (NHMRC ; grants 628331 , 023294 , and 510488 to Stephen J Kent). The funding source had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
3. Results
3.1. NK Cell Activating Antibodies to M1 and NP are Common in Influenza-exposed Healthy Adults
Influenza NP and M1 proteins are common targets for T cell immunity but whether they also elicit functionally important Abs that activate NK cells has not been well studied. Plasma from 14 healthy influenza-exposed human adults and four influenza-naïve pigtail macaques were first tested for NK cell activating Abs to M1 from the A/Puerto Rico/8/1934 (H1N1) influenza virus and NP from the H1N1 pdm09 influenza virus. The ability of plasma Abs to bind immobilized influenza proteins and activate primary NK cells to express the antiviral cytokine IFNγ and degranulation marker CD107a was studied by flow cytometry (Fig. 1 a).
Degranulation of NK cells would likely be more efficient in the presence of influenza-infected target cells, but for the purpose of studying NK cell activation by polyclonal Abs to individual influenza proteins in human sera or pooled IgG immobilized influenza proteins were used. Two additional influenza HA proteins from recently circulating strains, the HA of the A/Perth/19/2009 (H3N2) virus (H3 Perth09) and the HA of the H1N1 pdm09 virus (H1pdm09), were also tested for Ab-dependent NK cell activation (Fig. 1 b, c) since HA is a well-characterized target of ADCC (Jegaskanda et al., 2013a , Jegaskanda et al., 2013b , Jegaskanda et al., 2013c , Jegaskanda et al., 2013d , Jegaskanda et al., 2014a and Jegaskanda et al., 2014b ). An irrelevant viral Ag, HIV-1 gp140, was used to coat negative control wells since all healthy donors were HIV negative. Primary NK cells from different human donors commonly demonstrate skewing towards either IFNγ or CD107a expression, and the plasma or IgG samples tested can further bias the NK cell activation profile. IVIG (Fig. 1 a) appeared to elicit less CD107a expression in the primary NK cells we used compared to the responses observed in healthy donor plasma (Fig. 1 c). This is not surprising as we have previously shown that isolation of IgG from sera decreased CD107a expression in primary NK cells suggesting that soluble factors in plasma or sera, other than IgG, can alter NK cell activation profiles (Wren et al., 2012 ).
Plasma from influenza-exposed human adults induced substantial expression of IFNγ and CD107a in primary NK cells in response to both internal influenza proteins M1 and NP (Fig. 1 b, c). Following incubation with healthy donor plasma and M1, the median percentage of NK cell activation was 4.46% for IFNγ+ and 0.84% for CD107a+ compared to low levels of NK cell activation induced by influenza infection-naïve macaque plasma (median IFNγ+ : 0.6% and median CD107a+ : 0.37%, p < 0.001). Similarly, the proportion of NK cells expressing IFNγ and CD107a in response to anti-NP Abs was significantly greater in the presence of human donor plasma than in the presence naïve macaque plasma (median IFNγ+ : human plasma 2.72% vs macaque plasma 0.11%, p < 0.001; median CD107a+ : human plasma 1.58% vs macaque plasma 0.34%, p < 0.001). Influenza-exposed human plasma also led to more robust NK cell activation when PBMCs were stimulated with influenza Ags compared to HIV-1 gp140 stimulated cells (Fig. 1 b, c). The gp140-coated wells had very low levels of NK cell activation in the presence of healthy donor plasma (median IFNγ+ : 0.13% and median CD107a+ : 0.43%) confirming that activation was influenza specific and there were no detectable NK cell activating Abs against a viral Ag to which there has been no previous exposure. Unsurprisingly, HA proteins from both H1N1 pdm09 virus and A/Perth/19/2009 (H3N2) virus elicited high levels of NK cell activation (H1pdm09 median IFNγ+ : 5.24% and CD107a+ : 1.55%; H3 Perth09 median IFNγ+ : 12.01% and CD107a+ : 2.71%). For both HAs, the 14 healthy donor plasma samples had detectable NK cell activation (at least three times the percentage of IFNγ+ NK cells in gp140 coated wells with plasma). Remarkably M1 and NP also showed above background NK cell activation for all healthy donors tested. Thus, NK cell activating Abs specific for influenza M1 and NP are present in the plasma of healthy adults. To show that influenza proteins opsonised by polyclonal Abs are capable of activating NK cells without the involvement of other cells types we purified primary NK cells and performed an NK cell activation assay. When healthy donor plasma was added to surface (H1pdm09 and H3 Perth09) or internal (M1 and NP) influenza proteins, purified NK cells exhibited robust expression of IFNγ and CD107a (Supplementary Fig. 1 ) suggesting a direct mechanism of NK cell activation.
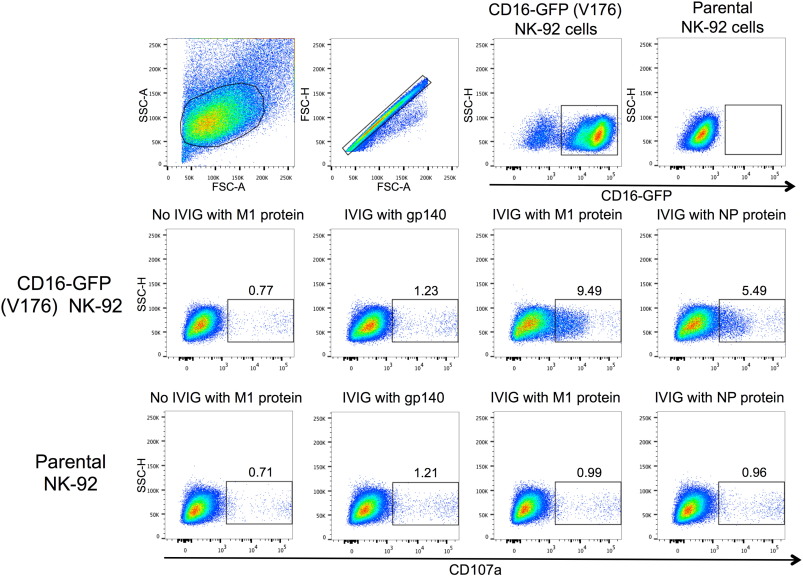
Primary NK cells in PBMCs exhibit variable levels of responsiveness to NK cell activating Abs over time and between donors. To confirm the above results we utilized an NK cell line, GFP-CD16 (V176) NK-92, as an alternative means for measuring NK cell activation. In addition, the larger numbers of NK-92 cells available allowed us to titrate M1 and NP-specific NK cell activating Abs in our cohort of 14 healthy donors (Fig. 2 b, d). NK-92 activation was assessed by surface expression of CD107a (Fig. 2 a). Ab-induced NK-92 activation to M1 protein was detected for all 14 healthy donors at a 1:10 plasma dilution (Fig. 2 b). Primary NK cell activation with undiluted plasma correlated with NK-92 activation for all plasma dilutions between 1:10 and 1:1280 for the M1 protein (correlation for 1:40 dilution shown in Fig. 2 c). All plasma samples also had detectable NK-92 activation to NP at a 1:10 dilution, but a slight prozone effect was observed for half of the plasma samples (Fig. 2 d). We speculate that high levels of NP-specific NK cell activating Abs may be inhibitory to NK-92 cells, yet there was still a significant correlation between primary NK cell activation with undiluted plasma and NK-92 activation for plasma dilutions between 1:10 and 1:10,240 (correlation for 1:40 dilution shown in Fig. 2 e). IFNγ was the marker of primary NK cell activation used for the correlations shown in Fig. 2 , however these analyses were also performed with percentages of primary NK cells expressing CD107a and yielded similar results (data not shown). To assess whether NK-92 activation was specifically mediated through FcγRIIIa (CD16a) we also performed an NK cell activation assay with the parental NK-92 cell line, which does not express FcγRIIIa. These results showed that parental NK-92 cells could not be activated in the presence of pooled human IgG and internal influenza proteins, while NK-92 cells transduced with FcγRIIIa (CD16a) demonstrated robust activation by CD107a expression (Supplementary Fig. 2 ). The strong correlation of Ab-mediated NK-92 cell activation with primary NK cell activation, as well as the FcγRIIIa dependence of their CD107a expression, suggests that NK-92 cells are a reasonable model of primary NK cell activation when studying influenza-specific Abs in humans.
3.2. NP and M1 Opsonised With Antibodies From Healthy Influenza-exposed Adults Bind Dimeric rsFcγRIIIa
ADCC Abs bind targets via their Fab regions and subsequently their Fc regions bind and crosslink FcγRIIIa to activate NK cells. A recently described dimeric rsFcγRIIIa binding ELISA (unpublished data; Wines et al.) was used to examine a subset of nine healthy donors and one influenza-naïve pigtail macaque. At a 1:10 plasma dilution all nine healthy influenza-exposed adults had detectable M1 and NP-specific Abs which reacted with Ag and bound dimeric rsFcγRIIIa (Fig. 3 a). Detectable receptor binding was defined as an OD value twice that of the corresponding control well with plasma but no influenza protein. The influenza-naïve macaque did not demonstrate detectable dimeric rsFcγRIIIa binding to Ab opsonised to M1 or NP at any plasma dilution tested. Median dimeric rsFcγRIIIa (normalised OD) for the nine human donors at a 1:10 plasma dilution was 0.29 for M1-specific Abs and 1.06 for NP-specific Abs. Abs opsonising M1 appeared to titre out rapidly for dimeric rsFcγRIIIa with all but one donor falling below the threshold of detection by the 1:320 plasma dilution. In contrast, most donors (7 out of 9) had Abs that opsonised NP with detectable dimeric rsFcγRIIIa binding at a 1:320 plasma dilution (Fig. 3 a).
The dimeric rsFcγRIIIa binding activity of M1 and NP opsonised with Abs correlated with primary NK cell activation. For the M1 protein, opsonisation with plasma diluted at 1:10 (Fig. 3 b; r = 0.89, p = 0.001), 1:32 (data not shown; r = 0.87, p = 0.002) and 1:100 (data not shown; r = 0.84, p = 0.004) resulted in dimeric rsFcγRIIIa binding activity that correlated significantly with primary NK cell activation. At a 1:320 dilution Ab opsonisation of M1 appeared to have decreased below the threshold of detectable dimeric rsFcγRIIIa binding (data not shown; r = 0.47, p = 0.17). Significant correlations between primary NK cell activation and dimeric rsFcγRIIIa binding with plasma dilutions of 1:32 (data not shown; r = 0.67, p = 0.04), 1:100 (data not shown; r = 0.72, p = 0.02) and 1:320 (Fig. 3 c; r = 0.72, p = 0.02) were also demonstrated for NP. At the 1:10 plasma dilution high levels of anti-NP Abs gave uniform, elevated dimeric rsFcγRIIIa binding across all healthy influenza-exposed donors and did not show a statistically significant correlation (data not shown; r = 0.48, p = 0.17). This highlights the importance of optimizing plasma dilutions for the dimeric rsFcγRIIIa binding ELISA based on the cohort and the Ag being tested. Collectively these results suggest that Abs capable of opsonising M1 and NP to form complexes with dimeric rsFcγRIIIa are present in most healthy donors and that this rsFcγRIIIa binding activity correlates with NK cell activation.
3.3. NK Cell Activation by Antibodies to M1 and NP in IVIG Samples
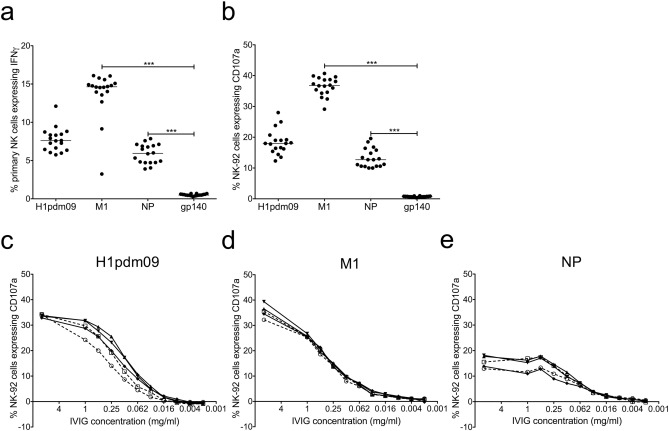
To further investigate the levels of NK cell activating Abs to internal influenza proteins across large numbers of subjects, the activation of primary NK and NK-92 cells was examined for 18 IVIG samples. Eight of the IVIG preparations were made with samples donated prior to the 2009 influenza pandemic (pre-2009) and ten were made with samples collected after the 2009 pandemic (post-2009). All these commercial IVIGs were prepared by pooling IgG samples from thousands of HIV-negative donors, allowing average influenza-specific NK cell activating Ab levels to be studied in the population. IVIG incubated with influenza proteins demonstrated robust activation of both primary NK cells (Fig. 4 a) and NK-92 cells (Fig. 4 b). The control HIV-1 gp140 protein induced only background levels of IVIG-mediated NK cell activation. The internal influenza protein M1 demonstrated robust activation of primary NK cells (median IFNγ+ : M1 14.6% vs gp140 0.48%, p < 0.0001) and NK-92 cells (median CD107a+ : M1 PR8 36.8% vs gp140 0.70%, p < 0.0001). NP also elicited significant levels of Ab-mediated activation of NK cells (median IFNγ+ : NP 5.95% vs gp140 0.48%, p < 0.0001) and NK-92 cells (median CD107a+ : NP 12.7% vs gp140 0.70%, p < 0.0001). The levels of NK cell activation clustered quite closely across the 18 IVIG samples for each influenza Ag tested (Fig. 4 a, b), this is not surprising due to the large number of donors used to generate each IVIG preparation. NK cell activation detected with IVIG demonstrates that M1 and NP-specific NK cell activating Abs are present in the population as a whole, which is consistent with what we observed in 14 healthy adult donors. There was no significant difference in primary NK cell or NK-92 activation between pre-2009 and post-2009 samples for the H1pdm09 protein as previously described (Jegaskanda et al., 2014a ), M1 or NP (data not shown), suggesting the advent of the 2009 pandemic did not have a major impact on population level ADCC responses to these influenza proteins.
|
|
|
Fig. 4. Influenza-specific NK cell activation by IVIG preparations and titration of H1pdm09, M1 and NP NK cell activating Abs in five IVIG samples. 18 IVIG preparations (10 mg/ml) were tested for primary NK cell (a) and NK-92 cell (b) activation to influenza proteins H1pdm09, M1 and NP as well as the irrelevant HIV-1 protein gp140. Values are unsubtracted with gp140 background shown for all samples. A Friedman test followed by a Wilcoxon matched pairs signed-rank test with a Bonferroni correction for multiple comparisons was used to compare influenza-specific NK activation by IVIGs to activation by gp140, a corrected p value of < 0.0167 considered significant. Titrations of NK cell activating Abs to H1pdm09 (c), M1 (d) and NP (e) were performed in a series of 2-fold dilutions for five IVIG samples two prepared pre-2009 (broken line, open symbol) and three prepared post-2009 (solid line, closed symbol). For all titrations (c–e) values were background subtracted with wells containing IVIG and gp140. *** = p < 0.001. |
The portion of activated NK cells at a single high concentration of IVIG (10 mg/ml) may not reflect the amount of NK cell activating Abs present. To compare the quantity of H1pdm09, M1 and NP-responsive NK cell activating Abs in the population five IVIG samples (two pre-2009 and three post-2009) were titrated in a series of two-fold dilutions with NK-92 cells. All five IVIG preparations showed similar titration patterns for each influenza protein (Fig. 4 c–e). NK cell activating Abs to HA, M1 and NP appeared to titre out at similar IVIG concentrations despite varying levels of NK-92 activation at the initial concentration of 10 mg/ml.
3.4. NK Cell Activation by Antibodies in Naturally Influenza-infected Adults
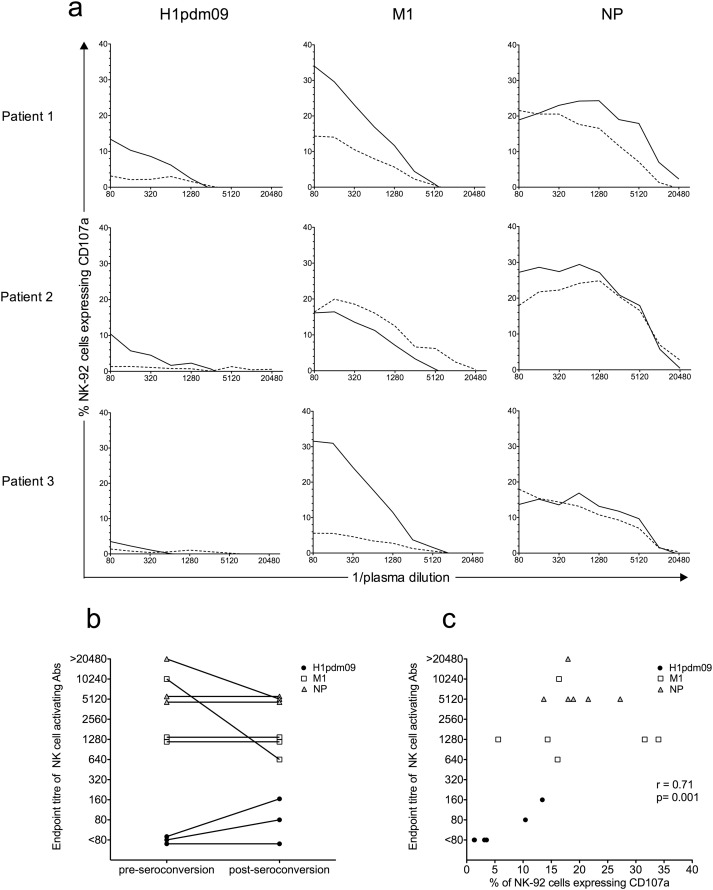
The above studies reflect NK cell activating Ab levels from individuals with unknown histories of influenza infections. To determine the impact of symptomatic influenza infection on the function and levels of human NK cell activating Abs, sequential sera samples ranging from pre to post-seroconversion were obtained from three adults and tested for NK cell activating Abs to HA, M1 and NP. We defined the first sample obtained from the subjects upon presentation with symptomatic illness as the “pre-seroconversion sample” although the acquisition of infection would have been prior to this time. The post-seroconversion samples were taken 12–17 days later. Post-seroconversion sera from all patients demonstrated increased HI titre and complement fixation to HA of the H1N1 pdm09 strain (Table 1 ). Titrations of patient sera were performed to compare the magnitude of NK cell activation and the levels of NK cell activating Abs between pre and post-seroconversion samples. At a 1:80 serum dilution all three patients showed a rise in NK-92 activation to the H1pdm09 protein post-seroconversion (post 3.5–14% CD107a+ vs pre 1–3% CD107a+ ). Patients 1 and 3 also showed a substantial rise in NK-92 activation to M1 protein in post-seroconversion sera (34–36% CD107a+ ) compared to pre-seroconversion samples (5–14% CD107a+ , Fig. 5 a). Only patient 2 showed some increase in NK-92 activation to NP after seroconversion at a 1:80 dilution (Fig. 5 a). Over most dilutions, there was higher NK cell activation in the post-seroconversion sample compared to the pre-seroconversion sample for both M1 and NP internal proteins as well as the H1pdm09 protein. Two of the three patients demonstrated a higher endpoint titre of NK cell activating Abs to HA post-seroconversion (patient 1: post 160 vs pre < 80; patient 2: post 80 vs pre < 80), whereas patient 3 showed no increase in endpoint titre post-seroconversion. Interestingly, the endpoint titres of M1 and NP-specific NK cell activating Abs remained constant pre and post-seroconversion for patients 1 and 3 (M1: 1280 and NP: 5120) and decreased post-seroconversion for patient 2 (M1: post 640 vs pre 10,240; NP: post 5120 vs pre > 20,480, Fig. 5 b). This suggests that although the potency of the Abs' ability to activate NK cells was enhanced, the amount of NK cell activating Abs was not increased. A strong correlation between endpoint titre of NK cell activating Abs and NK cell activation at a single dilution was observed when the three influenza Ags tested (HA, M1 and NP) were analysed together for pre and post-seroconversion sera samples (Fig. 5 c). This suggests that despite the lack of an increase in endpoint titre to M1 and NP post-seroconversion, there appeared to be significant relationship between NK-activating Ab levels and the degree of NK cell activation for these subjects with naturally acquired influenza infection.
|
|
|
Fig. 5. NK-92 activation by pre- and post-seroconversion sera samples from three naturally influenza-infected patients. a) A titration of NK-92 cell activating Abs, measured by percentage of CD107a+ cells, was performed with sera from subjects naturally infected with a suspected A/California/07/2009 (H1N1)-like influenza virus. NK cell activating Abs to influenza H1pdm09, M1 and NP were titrated in pre (broken line) and post-seroconversion (solid line) sera by 2-fold serial dilutions starting at a 1:80 dilution. All values were background subtracted with wells containing patient sera and gp140. b) Endpoint titres of NK cell activating Abs to H1pdm09 (closed black circles), M1 (open squares) and NP (grey triangles) are shown in pre- and post-seroconversion sera for the three subjects. Endpoint titres of NK cell activating Abs were defined as the last serum dilution before the percentage of NK-92 cells expressing CD107a fell below the threshold of three times background (calculated with wells containing sera and gp140). c) Correlation of NK cell activation (by CD107a expression) at a 1:80 serum dilution to endpoint titre of NK cell activating Abs. The percentage of CD107a+ NK-92 cells and the endpoint titres of NK cell activating Abs are depicted for all influenza proteins tested (H1pdm09, M1 and NP) with pre- and post-seroconversion sera from the three subjects naturally infected with influenza virus. Spearman correlation was used to compare NK-92 activation and endpoint titre of NK cell activating Abs, where p < 0.05 was considered significant. |
3.5. NK-92 Activation by Antibodies in Experimentally Influenza-infected Adults
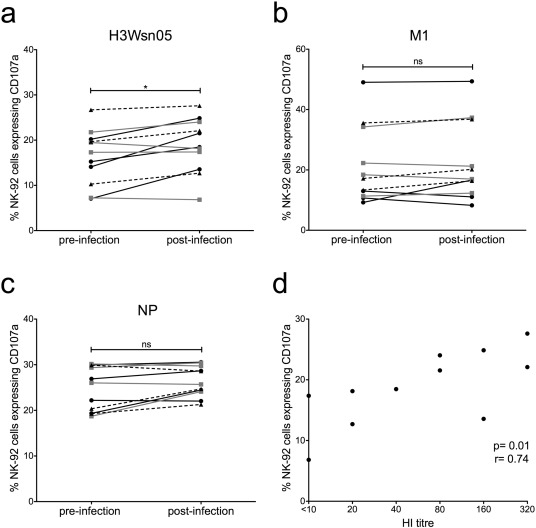
The precise timing of the influenza infection in the above study was not known, with the initial sample taken at first medical contact following influenza-like symptoms. To study anti-influenza NK cell activating Abs in a more controlled setting we assayed plasma samples from an experimental influenza infection where the pre-infection sample was taken prior to infection. The subjects studied were a cohort of 11 healthy volunteers infected with the influenza A/Wisconsin/67/2005 (H3N2) virus as controls for a previously published T cell based influenza vaccine study. The clinical and serological outcome varied widely among the volunteers (Lillie et al., 2012 ). Most but not all individuals (7 of 11) seroconverted to the challenge strain (HI > 40) at 36 days post-infection, but only five participants had detectable virus shedding (Table 2 ). Volunteers were seronegative (HI < 10) to the HA of the A/Wisconsin/67/2005 (H3N2) virus (H3 Wsn05) prior to infection, yet all pre-infection samples gave detectable baseline Ab-dependant NK-92 activation (7–26.7% CD107a+ ) to the challenge HA protein (Fig. 6 a). This is consistent with our prior work showing significant cross-reactive HA-specific NK cell activation in the absence of HI Abs (Jegaskanda et al., 2013a ). The conserved internal proteins M1 and NP also elicited detectable NK-92 activation in all donors prior to the experimental influenza infection (M1: 9.2%–49.1% CD107a+ ; NP: 18.7–30% CD107a+ ) (Fig. 6 b, c), consistent with the experiments above on healthy adults. NK-92 activation to the control HIV-1 gp140 protein was negligible. Pre-infection levels of NK cell activating Abs to the HA of the infecting virus and M1 varied substantially between individuals however no clear relationship between baseline NK-92 activation and disease severity, viral shedding or seroconversion was apparent in this small cohort.
|
|
|
Fig. 6. Influenza-specific Ab-dependent NK-92 activation in subjects experimentally infected with influenza and correlation with NAbs. The percentage of CD107a+ NK-92 cells following incubation with H3 Wsn05 (a), M1 (b) and NP (c) was compared for pre and post (36 days) infection plasma samples at a 1:20 dilution from 11 subjects experimentally infected with A/Wisconsin/67/2005 (H3N2) influenza virus. Black lines (circles) represent individuals that developed moderate/severe disease, grey lines (squares) depict subjects who showed mild disease and broken lines (triangles) are individuals that did not demonstrate any disease symptoms. All values were background subtracted with wells containing patient sera and gp140. Wilcoxon matched pairs signed-rank test was used to test for significant differences between pre- and post-infection plasma samples where p < 0.05 was considered significant. * = p < 0.05, ns = not significant. d) Correlation of HI titre to A/Wisconsin/67/2005 (H3N2) and percentage of NK-92 cells expressing CD107a for post-infection plasma samples. Spearman correlation was used to compare HI titre and NK-92 activation where p < 0.05 was considered significant. |
Across all 11 subjects, there was a significant rise in Ab-dependent NK-92 activation to the HA protein of the challenge virus between pre- and post-infection samples (p = 0.0137, Fig. 6 a) with a median percent change of 12.4% (Table 2 ). No significant difference was observed in NK-92 activation to M1 or NP between the pre- and post-infection samples (Fig. 6 b, c), with median percent changes of 3.4% and 3.7% respectively (Table 2 ). All four individuals who developed moderate/severe illness showed increased NK-92 activation to the HA of the infecting virus post-infection however, this pattern was not observed for M1 or NP (shown in black lines in Fig. 6 a–c). Full titrations of pre- and post-infection samples to HA, M1 and NP were not performed with the NK-92 activation assay for the 11 experimentally infected subjects, although the levels of NK cell activation at the 1:20 dilution studied correlated with endpoint titrations using the FcγRIIIa dimer binding assay above (data not shown). A significant correlation was observed between HI titre and NK-92 activation to the HA of the challenge virus in plasma samples taken 36 days post-infection (p = 0.01, r = 0.74), suggesting that infection with the A/Wisconsin/67/2005 (H3N2) virus is generating both HA-specific neutralising and NK cell activating Abs (Fig. 6 d).
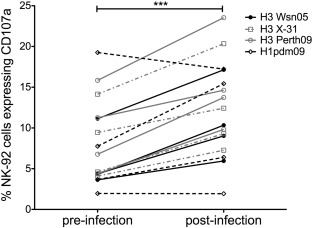
The four volunteers who developed moderate/severe disease and demonstrated greater NK-92 activation post-infection were examined for breadth of ADCC response to heterologous HAs. Following infection, all four individuals showed increased NK-92 activation to two heterologous H3 Ags tested (from the non-circulating X-31 virus and from the recently circulating A/Perth/19/2009 virus, Fig. 7 ). Two of the four individuals also showed increased ADCC response to the heterosubtypic HA of the H1N1 pdm09 virus (Fig. 7 ). This indicates that the NK cell activating Abs generated following symptomatic human influenza infection can recognize antigenically diverse, homosubtypic HAs and in some cases the HAs from heterosubtypic influenza viruses.
|
|
|
Fig. 7. Breadth of NK cell activating Abs to homosubtypic and heterosubtypic HA proteins in experimentally influenza-infected subjects with moderate/severe disease. NK-92 activation following stimulation with H3 Wsn05 (unbroken black line, closed circle), H3 X-31 (broken grey line, open square), H3 Perth09 (unbroken grey line, open circle) and H1pdm09 (broken black line, open diamond) was compared for pre and post (36 days) infection plasma samples at a 1:20 dilution from four subjects that demonstrated moderate/severe disease following A/Wisconsin/67/2005 (H3N2) infection. All values were background subtracted with wells containing patient sera and gp140. Wilcoxon matched pairs signed- rank test was used to test for significant differences between pre- and post-infection plasma samples where p < 0.05 was considered significant. *** = p < 0.001. |
3.6. ADCC Mediated Killing of Target Cells Expressing HA but Not M1 or NP
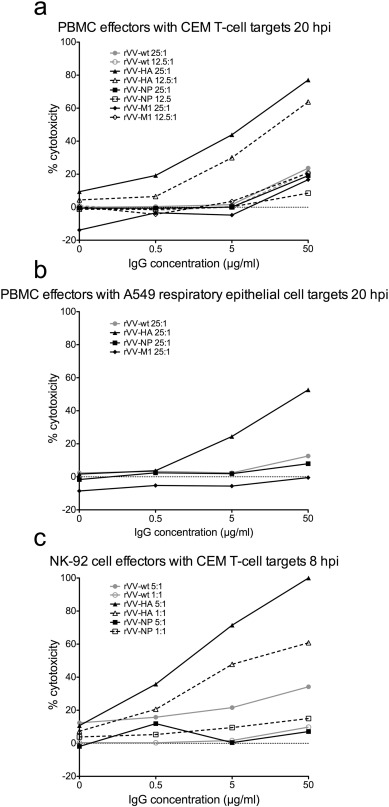
Although we found strong evidence that NP and M1-specific Abs were able to opsonise these Ags, bind dimeric rsFcγRIIIa and mediate NK cell activation, evaluating whether these Abs are able to recognize influenza-infected cells is difficult. Immune plasma contains Abs of different specificities (e.g. HA and NA) to which ADCC of influenza-infected will be directed (Jegaskanda et al., 2013a , Jegaskanda et al., 2013b , Jegaskanda et al., 2013c , Jegaskanda et al., 2013d and Jegaskanda et al., 2014a ). As a surrogate for influenza-infected cells we infected target cells with recombinant vaccinia viruses exclusively expressing either HA, NP or M1 Ags. Effector cells mediated robust Ab-dependent killing of CEM cells (Fig. 8 a, c) and A549 cells (Fig. 8 b) infected with the rVV expressing influenza HA in the presence of healthy adult IgG in all conditions tested. Low levels of ADCC were observed against CEM (Fig. 8 a, c) and A549 cells (Fig. 8 b) infected with the control rVV-wt particularly at the highest concentration of IgG (50 μg/ml). However even at an IgG concentration of 50 μg/ml, ADCC mediated killing of cells infected with rVV-HA was still 3 to 4-fold greater than that observed for cells infected with rVV-wt. Target cells infected with rVVs expressing influenza NP and M1 (Fig. 8 a–c) did not show enhanced cytotoxicity compared to cells infected with rVV-wt.
|
|
|
Fig. 8. ADCC mediated killing of targets cells infected with rVVs expressing individual influenza proteins. a) Percent cytotoxicity (measured by LDH release) of rVV infected CEM T-cell targets by PBMC effectors in the presence of 50 μg/ml, 5 μg/ml, 0.5 μg/ml or 0 μg/ml IgG from a healthy influenza-exposed adult. Effector PBMCs were added to targets 20 hour post-infection (hpi) with rVV at an effector to target ratio of 25:1 (solid lines, closed symbols) or 12.5:1 (broken lines, open symbols). b) Percent cytotoxicity of rVV infected lung epithelial A549 targets by effector PBMCs in the presence of healthy influenza-exposed donor IgG at concentrations of 50 μg/ml, 5 μg/ml, 0.5 μg/ml or 0 μg/ml. PBMC effectors were added to targets 20 hpi with rVV at an effector to target ratio of 25:1. c) Percent cytotoxicity of rVV infected CEM T-cell targets by NK-92 effector cells in the presence of healthy influenza-exposed donor IgG at 50 μg/ml, 5 μg/ml, 0.5 μg/ml or 0 μg/ml. Effector NK-92 cells were added to targets 8 hpi with rVV at effector to target ratios of 5:1 (solid lines, closed symbols) and 1:1 (broken lines, open symbols). |
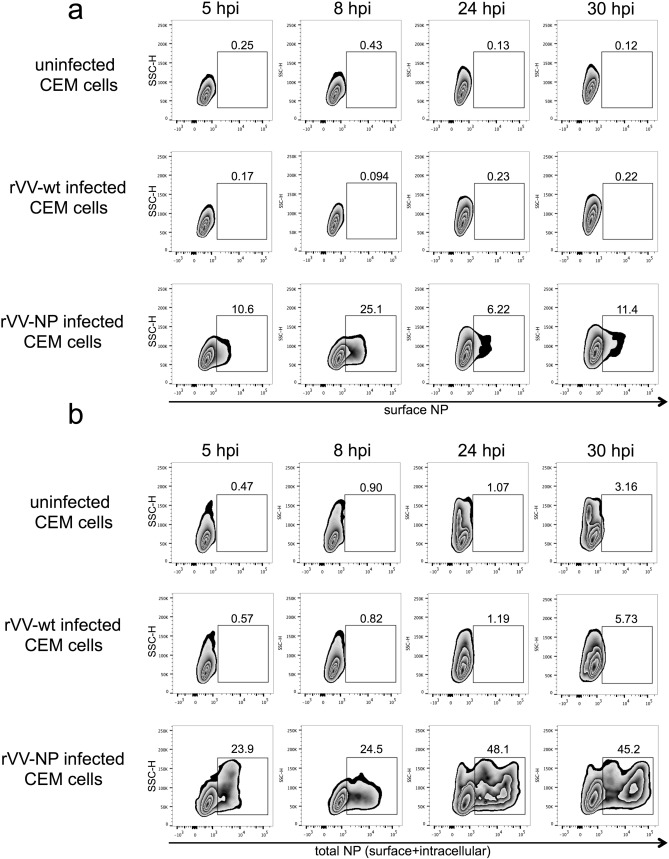
To establish that internal influenza proteins were present on the surface of cells in the rVV infection system, we infected CEM cells with rVV-NP or rVV-wt and surface stained with an anti-NP Ab conjugated to FITC (Abcam, Cambridge, UK). Surface NP expression was detectable at 5 hours post-infection (hpi) and peaked at 8 hpi with approximately 25% of unpermeabilized CEM cells showing surface NP by flow cytometry (Supplementary Fig. 3 ). At later time points of 24 hpi and 30 hpi with rVV-NP, surface NP declined to 6–11% of cells (Supplementary Fig. 3 ). In contrast, total surface and intracellular NP, measured in permeabilized rVV-NP infected CEM cells, increased between 5 hpi and 24 hpi with rVV-NP (Supplementary Fig. 3 ) showing a disparity between surface NP and total cellular NP in rVV-NP infected cells. There was no detectable surface NP on uninfected cells or CEM cells infected with rVV-wt (Supplementary Fig. 3 ). We then performed an ADCC killing assay using NK-92 effectors and CEM targets infected with rVV-wt, rVV-HA or rVV-NP at 8 hpi, which was the peak of surface NP detection by flow cytometry. At 8 hpi, rVV-HA infected cells elicited strong Ab-dependent killing by NK-92s (100% cytotoxicity) but consistent with previous assays at 20 h post-infection, ADCC was not increased with rVV-NP infected target cells (Fig. 8 c). In an rVV infection model, it appears that viruses expressing influenza HA alone, but not those expressing internal influenza proteins NP or M1, can mediate ADCC of infected target cells.
4. Discussion
Highly conserved influenza proteins have the potential to generate cross protective immunity and may provide the basis for a universal vaccine against influenza. It has long been known that influenza-exposed humans have Abs to NP (Ennis et al., 1982 , Sukeno et al., 1979 and Yamane et al., 1981 ), yet the functional consequences of humoral immunity to internal influenza Ags remain largely uncharacterised. In this study we have shown for the first time that individual and pooled human sera has opsonising Abs to internal influenza proteins capable of binding dimeric rsFcγRIIIa and activating NK cells.
Humans have been serially exposed to different influenza viruses from early childhood resulting in detectable baseline levels of influenza-specific ADCC Abs in the absence of NAbs (Terajima et al., 2015 ), but the impact of recent influenza infection on Ab-mediated human NK cell responses had not been previously explored. Acute influenza infection has been shown to decrease absolute numbers of peripheral blood NK cells and lower plasma levels of inflammatory cytokines (Jost et al., 2011 ). Inactivated influenza virions and HA can also inhibit primary NK cell activity in vitro (Mao et al., 2010 ). A limitation of this study was that PBMCs from the influenza-infected donors were not available, so NK cell dysfunction resulting from the influenza infections could not be assessed. Future studies could concomitantly study the response of autologous NK cells to functional influenza-specific Abs. We found that plasma from naturally and laboratory influenza-infected adults showed increased Ab-mediated NK cell activation to the HA protein of the infecting strain. The experimentally infected volunteers with the most severe disease symptoms also showed the largest post-infection rise in NK activation to the HA of the challenge strain. Plasma samples from subjects with moderate/severe symptoms following laboratory influenza challenge also demonstrated increased NK-92 activation to the HAs of two homosubtypic viruses post-infection. This suggests that symptomatic human influenza infection can induce cross-reactive NK cell activating Abs that recognize antigenically distinct viruses and potentially promote heterologous immunity, as we previously described in macaques (Jegaskanda et al., 2013c ).
Natural symptomatic but not experimental influenza infection increased Ab-mediated NK cell activation to the internal protein M1 and to a lesser extent NP. Donor infection history, strain/subtype of infecting virus, route of infection and infectious dose may affect the capacity of a given influenza infection to boost Ab-dependent NK cell activation. During natural influenza infection Ab-dependent NK cell activation was increased to the HA of the infecting virus and M1 following seroconversion. Surprisingly, when the NK cell activating Abs to HA and M1 were serially diluted it was revealed that in some cases the endpoint titres were identical in pre- and post-seroconversion samples. These results suggest that the total levels of NK cell activating Abs were not increased post-seroconversion but that the Abs induced or expanded after seroconversion are more potent activators of NK cells. This could be due to a number of factors including enhanced Ab avidity for its Ag or altered glycosylation of the IgG Fc region, which can change the binding affinity for Fc receptors (Ferrara et al., 2006 , Ferrara et al., 2011 and Shinkawa et al., 2003 ) and lead to altered immune effector functions like ADCC. Differential Fc glycosylation or increased Ab avidity post-seroconversion may explain the observation that NK cell activation at a single IgG concentration is variable in samples that demonstrate similar quantities of NK cell activating Abs by titration.
The role of NK cell activating Abs to internal influenza proteins in the control of influenza infection is controversial. Animal models of influenza infection suggest that Abs to conserved Ags like M1 and NP are important for heterosubtypic immunity (LaMere et al., 2011b ). Likewise, passive transfer of immune serum from mice vaccinated with NP into naïve or B-cell deficient mice decreased influenza viral load (Carragher et al., 2008 ). Since Abs to internal influenza Ags cannot prevent viral entry by neutralisation (Gerhard et al., 1997 and Mozdzanowska et al., 1999 ), anti -NP IgG activity is likely mediated through complex interactions with effector cells ( LaMere et al., 2011b ). In B cell deficient mice the antiviral action of NP reactive Abs is Fc receptor dependent and significantly reduced in Fc receptor γ-common chain knockout (FcRγ−/− ) mice (LaMere et al., 2011b ). The nature of these antiviral effects and their relevance to human influenza infection is unclear. Specifically it is unknown if M1 and NP Abs activate effector cells that directly kill infected targets (ADCC) or act indirectly by cytokine-mediated stimulation of other antiviral cell types.
Indeed, this study found NK cell activating Abs to highly conserved influenza Ags, but we were not able to detect Ab-mediated killing of target cells infected with rVVs expressing either NP or M1. Internal influenza proteins are present in the extracellular environment following their release from dead or infected cells (LaMere et al., 2011b ) and NP is present on the surface of influenza-infected cells in vitro as early as one hour post-infection (Bodewes et al., 2013 , Virelizier et al., 1977 and Yewdell et al., 1981 ). In spite of a robust rVV infection system as evidenced by consistently high levels of ADCC against rVV-HA infected cells, we found that only a small percentage (up to ~ 25%) of target CEM cells infected with rVV-NP had NP on the cell surface, which may be insufficient for detection of Ab-mediated killing in the assays employed. Influenza and vaccinia viruses differ in genome composition, replication cycle and tissue tropism therefore it is not known if the rVV expression system accurately reflects an in vivo influenza infection. Influenza-infected cells have been shown to express relatively low levels of surface NP compared to surface glycoproteins HA and NA, which were approximately 10-fold more abundant (Yewdell et al., 1981 ). Future studies using influenza-infected cells as targets in Ab-mediated killing assays would be informative, but such assays are problematic since influenza-infected cells are susceptible to HA and NA-specific ADCC in polyclonal serum. To carry out Ab-mediated killing assays specifically targeting surface NP and M1 on influenza-infected cells would require monoclonal Abs (mAbs) with known ADCC activity. However most currently available mAbs are not isolated with their native Fc regions and glycosylation patterns, which can significantly impact ADCC functionality (Arnold et al., 2007 , Mahan et al., 2016 and Vidarsson et al., 2014 ).
Apart from opsonising infected cells the Abs may also form circulating immune complexes (ICs) by opsonising soluble M1 and NP. These circulating ICs may activate NK cells or other effectors to enhance immune responses. Innate immune cell activation by circulating ICs would lead to the release of pro-inflammatory cytokines and chemokines capable of activating and recruiting different effector cell types (macrophages, neutrophils and T-cells) as reported in the mouse infection model.
Collectively, influenza-exposed humans have detectable NK cell activating Abs to highly conserved internal influenza proteins. M1 and NP reactive Abs are capable of mediating robust NK cell activation and degranulation, enhancing the local immune response to influenza. Further, influenza infection boosts NK cell activating Abs to the HA of the infecting virus and induces cross-reactive Abs to the HAs of homosubtypic influenza viruses. We speculate that opsonising Abs to both surface and internal proteins are capable of activating NK cells or other innate Fc receptor expressing effectors to confer some level of heterologous influenza protection in humans.
The following are the supplementary data related to this article.
|
|
|
Supplementary Fig. 1. Influenza-specific NK cell activation by healthy donor plasma in primary and purified primary NK cells. Gating strategies are shown for primary NK cells in PBMCs (a) and purified primary NK cells (b). For both conditions, CD3 − CD56 + dim primary NK cells were selected for analysis using IFNγ and CD107a as activation markers. PBMCs or purified NK cells were incubated with influenza protein (600 ng/well) in the absence of plasma from a healthy influenza-exposed donor, irrelevant viral protein gp140 (600 ng/well) with plasma from an influenza-exposed donor and influenza proteins (M1 and NP) with plasma from an influenza-exposed donor. c) Primary and purified primary NK cell activation is shown with plasma from a healthy influenza-exposed donor (+) and an influenza-naïve pigtail macaque (−) by IFNγ and/or CD107a expression to HA of A/California/04/2009 (H1pdm09), HA of A/Perth/19/2009 (H3Perth09), M1 of A/Puerto Rico/8/1934 (M1), NP of A/California/07/2009 (NP) and irrelevant viral protein gp140. This assay showed similar results with a second healthy plasma donor and an IVIG preparation (10 mg/ml). |
|
|
|
Supplementary Fig. 2. Activation of GFP-CD16 (V176) NK-92 and parental NK-92 cells by M1- and NP-specific Abs in an IVIG preparation. GFP-CD16 (V176) NK-92 and parental NK-92 cells were gated for size and granularity (FSC-A vs SSC-A) ensuring single cells (FSC-A vs FSC-H). CD16-GFP expression was shown for the CD16-GFP (V176) NK-92 cells (top panel labelled “CD16-GFP (V176) NK-92”) and the parental NK-92 cells (top panel labelled “Parental NK-92”). Single CD16-GFP (V176) NK-92 (middle panel) and parental NK-92 (bottom panel) cells were analysed for activation by CD107a expression following incubation with or without IVIG (10 mg/ml) and influenza proteins (M1 and NP at 600 ng/well) or an irrelevant viral protein (gp140 at 600 ng/well). |
|
|
|
Supplementary Fig. 3. Surface and total expression of NP for rVV-NP infected CEM cells between 5 and 30 hours post-infection. Unpermeabilized (a) or permeabilized (b) CEM cells were stained with an anti-NP Ab conjugated to FITC and measured by flow cytometry at 5, 8, 24 and 30 hours post-infection (hpi) with no virus, rVV-wt or rVV-NP. |
Author Contributions
HAV performed research, data acquisition and interpretation. HAV and SJK drafted the manuscript. SJK, SJ, MSP and HAV were responsible for study concept and design. FASB and SJ provided extensive technical assistance with experimental work and assay set-up. SR, IB, KL, SCG and TL provided human samples and characterized the cohorts examined in this study. BW and PMH developed the FcγRIIIa (CD16a) dimer ELISA and gave expert guidance in the use of this assay. WC provided the rVV stocks and technical assistance with rVV infections. SJK received funding for this work. All authors contributed to critical revision of the manuscript.
Conflicts of Interest
SR is an employee of Seqirus Ltd (formerly bioCSL Ltd), a company that produces the seasonal influenza vaccine. SCG is a consultant to Vaccitech, a company which is developing an influenza vaccination. The remaining authors have no conflicts of interest.
Acknowledgements
We would like to sincerely thank all healthy volunteers and clinical subjects who donated blood samples for this research. We would also like to thank Drs Jonathan Yewdell and Jack Bennink from the NIH (Bethesda, MD) for the rVVs expressing individual influenza proteins. The WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health and Aging.
References
- Arnold et al., 2007 J.N. Arnold, M.R. Wormald, R.B. Sim, P.M. Rudd, R.A. Dwek; The impact of glycosylation on the biological function and structure of human immunoglobulins; Annu. Rev. Immunol., 25 (2007), pp. 21–50
- Atmar et al., 2007 R.L. Atmar, W.A. Keitel, T.R. Cate, F.M. Munoz, F. Ruben, R.B. Couch; A dose-response evaluation of inactivated influenza vaccine given intranasally and intramuscularly to healthy young adults; Vaccine, 25 (2007), pp. 5367–5373
- Bodewes et al., 2013 R. Bodewes, M.M. Geelhoed-Mieras, J. Wrammert, et al.; In vitro assessment of the immunological significance of a human monoclonal antibody directed to the influenza a virus nucleoprotein; Clin. Vaccine Immunol., 20 (2013), pp. 1333–1337
- Carragher et al., 2008 D.M. Carragher, D.A. Kaminski, A. Moquin, L. Hartson, T.D. Randall; A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus; J. Immunol., 181 (2008), pp. 4168–4176
- Chen et al., 2014 L. Chen, D. Zanker, K. Xiao, C. Wu, Q. Zou, W. Chen; Immunodominant CD4 + T-cell responses to influenza A virus in healthy individuals focus on matrix 1 and nucleoprotein; J. Virol., 88 (2014), pp. 11760–11773
- Cox, 1999 J.H. Cox; HIV-1-specific antibody-dependent cellular cytotoxicity (ADCC); Methods Mol. Med., 17 (1999), pp. 373–381
- DiLillo et al., 2014 D.J. DiLillo, G.S. Tan, P. Palese, J.V. Ravetch; Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo; Nat. Med., 20 (2014), pp. 143–151
- El Bakkouri et al., 2011 K. El Bakkouri, F. Descamps, M. De Filette, et al.; Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection; J. Immunol., 186 (2011), pp. 1022–1031
- Ellebedy et al., 2014 A.H. Ellebedy, F. Krammer, G.M. Li, et al.; Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans; Proc. Natl. Acad. Sci. U. S. A., 111 (2014), pp. 13133–13138
- Ennis et al., 1982 F.A. Ennis, Q. Yi-Hua, G.C. Schild; Antibody and cytotoxic T lymphocyte responses of humans to live and inactivated influenza vaccines; J. Gen. Virol., 58 (1982), pp. 273–281
- Epstein et al., 2005 S.L. Epstein, W.P. Kong, J.A. Misplon, et al.; Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein; Vaccine, 23 (2005), pp. 5404–5410
- Ferrara et al., 2006 C. Ferrara, P. Brunker, T. Suter, S. Moser, U. Puntener, P. Umana; Modulation of therapeutic antibody effector functions by glycosylation engineering: influence of Golgi enzyme localization domain and co-expression of heterologous beta1, 4-N-acetylglucosaminyltransferase III and Golgi alpha-mannosidase II; Biotechnol. Bioeng., 93 (2006), pp. 851–861
- Ferrara et al., 2011 C. Ferrara, S. Grau, C. Jager, et al.; Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose; Proc. Natl. Acad. Sci. U. S. A., 108 (2011), pp. 12669–12674
- Fielding et al., 2011 J.E. Fielding, K.A. Grant, K. Garcia, H.A. Kelly; Effectiveness of seasonal influenza vaccine against pandemic (H1N1) 2009 virus, Australia, 2010; Emerg. Infect. Dis., 17 (2011), pp. 1181–1187
- Gerhard et al., 1997 W. Gerhard, K. Mozdzanowska, M. Furchner, G. Washko, K. Maiese; Role of the B-cell response in recovery of mice from primary influenza virus infection; Immunol. Rev., 159 (1997), pp. 95–103
- Gong et al., 1994 J.H. Gong, G. Maki, H.G. Klingemann; Characterization of a human cell line (NK-92) with phenotypical and functional characteristics of activated natural killer cells; Leukemia, 8 (1994), pp. 652–658
- Gooneratne et al., 2015 S.L. Gooneratne, J. Richard, W.S. Lee, A. Finzi, S.J. Kent, M.S. Parsons; Slaying the Trojan horse: natural killer cells exhibit robust anti-HIV-1 antibody-dependent activation and cytolysis against allogeneic T cells; J. Virol., 89 (2015), pp. 97–109
- Greenberg et al., 1978 S.B. Greenberg, B.S. Criswell, H.R. Six, R.B. Couch; Lymphocyte cytotoxicity to influenza virus-infected cells: response to vaccination and virus infection; Infect. Immun., 20 (1978), pp. 640–645
- Hardelid et al., 2011 P. Hardelid, D.M. Fleming, J. McMenamin, et al.; Effectiveness of pandemic and seasonal influenza vaccine in preventing pandemic influenza A(H1N1)2009 infection in England and Scotland 2009–2010; Euro Surveill. Bull. Eur. Mal. Transmis. Eur. Commun. Dis. Bull., 16 (2011)
- Hashimoto et al., 1983a G. Hashimoto, P.F. Wright, D.T. Karzon; Antibody-dependent cell-mediated cytotoxicity against influenza virus-infected cells; J. Infect. Dis., 148 (1983), pp. 785–794
- Hashimoto et al., 1983b G. Hashimoto, P.F. Wright, D.T. Karzon; Ability of human cord blood lymphocytes to mediate antibody-dependent cellular cytotoxicity against influenza virus-infected cells; Infect. Immun., 42 (1983), pp. 214–218
- Hessel et al., 2014 A. Hessel, H. Savidis-Dacho, S. Coulibaly, et al.; MVA vectors expressing conserved influenza proteins protect mice against lethal challenge with H5N1, H9N2 and H7N1 viruses; PLoS One, 9 (2014), Article e88340
- Huber et al., 2001 V.C. Huber, J.M. Lynch, D.J. Bucher, J. Le, D.W. Metzger; Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections; J. Immunol., 166 (2001), pp. 7381–7388
- Jegaskanda et al., 2013a S. Jegaskanda, E.R. Job, M. Kramski, et al.; Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies; J. Immunol., 190 (2013), pp. 1837–1848
- Jegaskanda et al., 2013b S. Jegaskanda, K.L. Laurie, T.H. Amarasena, et al.; Age-associated cross-reactive antibody-dependent cellular cytotoxicity toward 2009 pandemic influenza A virus subtype H1N1; J. Infect. Dis., 208 (2013), pp. 1051–1061
- Jegaskanda et al., 2013c S. Jegaskanda, J.T. Weinfurter, T.C. Friedrich, S.J. Kent; Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques; J. Virol., 87 (2013), pp. 5512–5522
- Jegaskanda et al., 2013d S. Jegaskanda, T.H. Amarasena, K.L. Laurie, et al.; Standard trivalent influenza virus protein vaccination does not prime antibody-dependent cellular cytotoxicity in macaques; J. Virol., 87 (2013), pp. 13706–13718
- Jegaskanda et al., 2014a S. Jegaskanda, K. Vandenberg, K.L. Laurie, et al.; Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity in intravenous immunoglobulin as a potential therapeutic against emerging influenza viruses; J. Infect. Dis., 210 (2014), pp. 1811–1822
- Jegaskanda et al., 2014b S. Jegaskanda, P.C. Reading, S.J. Kent; Influenza-specific antibody-dependent cellular cytotoxicity: toward a universal influenza vaccine; J. Immunol., 193 (2014), pp. 469–475
- Jost et al., 2011 S. Jost, H. Quillay, J. Reardon, et al.; Changes in cytokine levels and NK cell activation associated with influenza; PLoS One, 6 (2011), Article e25060
- Kim et al., 2014 M.C. Kim, Y.N. Lee, H.S. Hwang, et al.; Influenza M2 virus-like particles confer a broader range of cross protection to the strain-specific pre-existing immunity; Vaccine, 32 (2014), pp. 5824–5831
- Krause et al., 2011 J.C. Krause, T. Tsibane, T.M. Tumpey, C.J. Huffman, C.F. Basler, J.E. Crowe Jr.; A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin; J. Virol., 85 (2011), pp. 10905–10908
- Lambe et al., 2013 T. Lambe, J.B. Carey, Y. Li, et al.; Immunity against heterosubtypic influenza virus induced by adenovirus and MVA expressing nucleoprotein and matrix protein-1; Sci. Rep., 3 (2013), p. 1443
- Lamere et al., 2011a M.W. Lamere, A. Moquin, F.E. Lee, et al.; Regulation of antinucleoprotein IgG by systemic vaccination and its effect on influenza virus clearance; J. Virol., 85 (2011), pp. 5027–5035
- LaMere et al., 2011b M.W. LaMere, H.T. Lam, A. Moquin, et al.; Contributions of antinucleoprotein IgG to heterosubtypic immunity against influenza virus; J. Immunol., 186 (2011), pp. 4331–4339
- Laoprasopwattana et al., 2007 K. Laoprasopwattana, D.H. Libraty, T.P. Endy, et al.; Antibody-dependent cellular cytotoxicity mediated by plasma obtained before secondary dengue virus infections: potential involvement in early control of viral replication; J. Infect. Dis., 195 (2007), pp. 1108–1116
- Lee et al., 2014 Y.N. Lee, Y.T. Lee, M.C. Kim, et al.; Fc receptor is not required for inducing antibodies but plays a critical role in conferring protection after influenza M2 vaccination; Immunology, 143 (2014), pp. 300–309
- Lillie et al., 2012 P.J. Lillie, T.K. Berthoud, T.J. Powell, et al.; Preliminary assessment of the efficacy of a T-cell-based influenza vaccine, MVA-NP + M1, in humans; Clin. Infect. Dis., 55 (2012), pp. 19–25
- Mackett et al., 1984 M. Mackett, G.L. Smith, B. Moss; General method for production and selection of infectious vaccinia virus recombinants expressing foreign genes; J. Virol., 49 (1984), pp. 857–864
- Mahan et al., 2016 A.E. Mahan, M.F. Jennewein, T. Suscovich, et al.; Antigen-specific antibody glycosylation is regulated via vaccination; PLoS Pathog., 12 (2016), Article e1005456
- Mao et al., 2010 H. Mao, W. Tu, Y. Liu, et al.; Inhibition of human natural killer cell activity by influenza virions and hemagglutinin; J. Virol., 84 (2010), pp. 4148–4157
- Margine et al., 2013a I. Margine, R. Hai, R.A. Albrecht, et al.; H3N2 influenza virus infection induces broadly reactive hemagglutinin stalk antibodies in humans and mice; J. Virol., 87 (2013), pp. 4728–4737
- Margine et al., 2013b I. Margine, F. Krammer, R. Hai, et al.; Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses; J. Virol., 87 (2013), pp. 10435–10446
- Martin Mdel et al., 2010 P. Martin Mdel, S. Seth, D.G. Koutsonanos, J. Jacob, R.W. Compans, I. Skountzou; Adjuvanted influenza vaccine administered intradermally elicits robust long-term immune responses that confer protection from lethal challenge; PLoS One, 5 (2010), Article e10897
- Mozdzanowska et al., 1999 K. Mozdzanowska, K. Maiese, M. Furchner, W. Gerhard; Treatment of influenza virus-infected SCID mice with nonneutralizing antibodies specific for the transmembrane proteins matrix 2 and neuraminidase reduces the pulmonary virus titer but fails to clear the infection; Virology, 254 (1999), pp. 138–146
- O'Brien et al., 2011 K.B. O'Brien, T.E. Morrison, D.Y. Dundore, M.T. Heise, S. Schultz-Cherry; A protective role for complement C3 protein during pandemic 2009 H1N1 and H5N1 influenza A virus infection; PLoS One, 6 (2011), Article e17377
- Ohta et al., 2011 R. Ohta, Y. Torii, M. Imai, H. Kimura, N. Okada, Y. Ito; Serum concentrations of complement anaphylatoxins and proinflammatory mediators in patients with 2009 H1N1 influenza; Microbiol. Immunol., 55 (2011), pp. 191–198
- Peiris et al., 2009 J.S. Peiris, W.W. Tu, H.L. Yen; A novel H1N1 virus causes the first pandemic of the 21st century; Eur. J. Immunol., 39 (2009), pp. 2946–2954
- Sandbulte et al., 2011 M.R. Sandbulte, K.B. Westgeest, J. Gao, et al.; Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses; Proc. Natl. Acad. Sci. U. S. A., 108 (2011), pp. 20748–20753
- Shil et al., 2011 P. Shil, S. Chavan, S. Cherian; Molecular basis of antigenic drift in Influenza A/H3N2 strains (1968–2007) in the light of antigenantibody interactions; Bioinformation, 6 (2011), pp. 266–270
- Shinkawa et al., 2003 T. Shinkawa, K. Nakamura, N. Yamane, et al.; The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity; J. Biol. Chem., 278 (2003), pp. 3466–3473
- Smith et al., 1987 G.L. Smith, J.Z. Levin, P. Palese, B. Moss; Synthesis and cellular location of the ten influenza polypeptides individually expressed by recombinant vaccinia viruses; Virology, 160 (1987), pp. 336–345
- Sukeno et al., 1979 N. Sukeno, Y. Otsuki, J. Konno, et al.; Anti-nucleoprotein antibody response in influenza A infection; Tohoku J. Exp. Med., 128 (1979), pp. 241–249
- Terajima et al., 2015 M. Terajima, M.D. Co, J. Cruz, F.A. Ennis; High antibody-dependent cellular cytotoxicity antibody titers to H5N1 and H7N9 avian influenza A viruses in healthy US adults and older children; J. Infect. Dis., 212 (2015), pp. 1052–1060
- Ulmer et al., 1993 J.B. Ulmer, J.J. Donnelly, S.E. Parker, et al.; Heterologous protection against influenza by injection of DNA encoding a viral protein; Science, 259 (1993), pp. 1745–1749
- Vella et al., 1980 S. Vella, G. Rocchi, S. Resta, M. Marcelli, A. De Felici; Antibody reactive in antibody-dependent cell-mediated cytotoxicity following influenza virus vaccination; J. Med. Virol., 6 (1980), pp. 203–211
- Vidarsson et al., 2014 G. Vidarsson, G. Dekkers, T. Rispens; IgG subclasses and allotypes: from structure to effector functions; Front. Immunol., 5 (2014), p. 520
- Virelizier et al., 1977 J.L. Virelizier, A.C. Allison, J.S. Oxford, G.C. Schild; Early presence of ribonucleoprotein antigen on surface of influenza virus-infected cells; Nature, 266 (1977), pp. 52–54
- Vogt et al., 2011 M.R. Vogt, K.A. Dowd, M. Engle, et al.; Poorly neutralizing cross-reactive antibodies against the fusion loop of West Nile virus envelope protein protect in vivo via Fcgamma receptor and complement-dependent effector mechanisms; J. Virol., 85 (2011), pp. 11567–11580
- Wang et al., 2010 T.T. Wang, G.S. Tan, R. Hai, et al.; Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins; PLoS Pathog., 6 (2010), Article e1000796
- World Health Organization, 2015 World Health Organization; Influenza (seasonal); Available at: http://www.who.int/mediacentre/factsheets/fs211/en/ (Accessed February 5th) (2015)
- Wraith et al., 1987 D.C. Wraith, A.E. Vessey, B.A. Askonas; Purified influenza virus nucleoprotein protects mice from lethal infection; J. Gen. Virol., 68 (Pt 2) (1987), pp. 433–440
- Wren et al., 2012 L. Wren, M.S. Parsons, G. Isitman, et al.; Influence of cytokines on HIV-specific antibody-dependent cellular cytotoxicity activation profile of natural killer cells; PLoS One, 7 (2012), Article e38580
- Xiao et al., 2010 P. Xiao, J. Zhao, L.J. Patterson, et al.; Multiple vaccine-elicited nonneutralizing antienvelope antibody activities contribute to protective efficacy by reducing both acute and chronic viremia following simian/human immunodeficiency virus SHIV89·6P challenge in rhesus macaques; J. Virol., 84 (2010), pp. 7161–7173
- Yamane et al., 1981 N. Yamane, T. Odagiri, J. Arikawa, N. Ishida; Reversed single-radial-immunodiffusion test: the method for the assay of the antibody to influenza A nucleoprotein; Tohoku J. Exp. Med., 133 (1981), pp. 245–255
- Yang et al., 2012 P. Yang, L. Zhang, W. Shi, et al.; Seroprevalence of pandemic (H1N1) 2009 influenza and effectiveness of 2010/2011 influenza vaccine during 2010/2011 season in Beijing, China; Influenza Other Respir. Viruses, 6 (2012), pp. 381–388
- Yewdell et al., 1981 J.W. Yewdell, E. Frank, W. Gerhard; Expression of influenza A virus internal antigens on the surface of infected P815 cells; J. Immunol., 126 (1981), pp. 1814–1819
- Yewdell et al., 1985 J.W. Yewdell, J.R. Bennink, G.L. Smith, B. Moss; Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes; Proc. Natl. Acad. Sci. U. S. A., 82 (1985), pp. 1785–1789
Document information
Published on 06/04/17
Licence: Other
Share this document
Keywords
claim authorship
Are you one of the authors of this document?